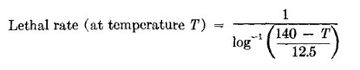Difference between revisions of "Quality Assurance"
(→Yeast Washing) |
|||
| (265 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | ''' | + | '''Quality Assurance''' refers to the process if developing standard operating procedures for proactively avoiding quality problems <ref name="Diffen">[https://www.diffen.com/difference/Quality_Assurance_vs_Quality_Control "Quality Assurance vs. Quality Control". Diffen website. Retrieved 03/28/2018.]</ref>. In the brewing industry, this includes avoiding off-flavors from contamination, dissolved oxygen in beer, fermentation and ingredient issues, etc. Although quality assurance in the brewing industry covers a wide range of approaches such as verifying yeast health and pitching rates, recording fermentation data, performance tracking, etc. (see the [[Quality_Assurance#External_Resources|External Resources]] below and [[Laboratory Techniques]]) <ref>Zachary Taggart. "Quality Assurance for Small Breweries: The Key to Growth". CBC 2018.</ref>, this article will focus primarily on avoiding microbial contamination through cleaning and disinfecting, particularly in the area of sharing a space for both pure culture fermentation and [[Mixed Fermentation|mixed fermentation]] beer. |
| − | ' | + | ==Overview== |
| + | Any microorganism that is introduced into a beer unintentionally and can survive in the beer is considered a "beer spoiler". Beer can be affected by contaminants via raw ingredients, poor sanitation, incorrect pasteurization, brewery environment air pollution, and inadequate ethanol production <ref>[https://www.mdpi.com/2304-8158/11/17/2693 Ciont, C.; Epuran, A.; Kerezsi, A.D.; Coldea, T.E.; Mudura, E.; Pasqualone, A.; Zhao, H.; Suharoschi, R.; Vriesekoop, F.; Pop, O.L. Beer Safety: New Challenges and Future Trends within Craft and Large-Scale Production. Foods 2022, 11, 2693. https://doi.org/10.3390/foods11172693.]</ref>. One survey of 38 craft beers in the Spanish market found that 68% of them had some presence of unwanted microbes, with beers under 5% ABV being more susceptible than higher ABV beers, indicating that craft breweries in particular may have a high degree of contamination issues <ref>[https://www.jmbfs.org/issue/june-july-2020-vol-9-no-6/jmbfs_2132_garcia-lopez/?issue_id=7366&article_id=25 CONTAMINANT MICROBIOTA IN CRAFT BEERS. Marta García López, Elena Rocheb, Encarnación Rodríguez. 2020.]</ref>. While most microorganisms cannot survive in beer due to the hops, low pH, alcohol content, relatively high carbon dioxide, and shortage of nutrients, certain species are considered to be beer spoilage organisms due to their ability to adapt to brewing conditions (namely hops, ethanol, and low pH) and sometimes form biofilms that help them resist cleaning. Some are able to survive in beer and make a potential impact on the beer's flavor by producing acidity, phenols, turbidity/ropiness via exopolysaccharides (EPS), and/or super-attenuation (which can cause gushing or in extreme cases exploding bottles/cans) with just a few surviving cells. These effects can sometimes manifest days or even weeks after packaging, and longer storage or non-refrigerated storage can increase the potential for beer spoilers to negatively impact the beer. Bacteria species that have adapted to the brewing environment tend to be hop tolerant, but strains of the same species found outside of breweries are not tolerant of brewing conditions. It is thought that these species evolved to carry the genes to adapt to brewing conditions during the 5th to 9th centuries when hops were first being used in brewing, and that this evolution gave them a specialized adaption to the brewing environment where few competitors can survive <ref name="Suzuki_2012">[https://onlinelibrary.wiley.com/doi/abs/10.1002/j.2050-0416.2011.tb00454.x 125th Anniversary Review: Microbiological Instability of Beer Caused by Spoilage Bacteria. Ken Suzuki. 2012. DOI: https://doi.org/10.1002/j.2050-0416.2011.tb00454.x]</ref>. Hop tolerant lactic acid bacteria have been found on the surfaces of many places in the brewing environment, including the fermentation area, bottling area, and cold storage. Hop tolerant lactic acid bacteria have been isolated from the air in at least one brewery in the fermentation and bottling areas <ref>[https://www.tandfonline.com/doi/abs/10.1094/ASBCJ-2017-4294-01?src=recsys Distribution of Lactobacillus and Pediococcus in a Brewery Environment. Jorge Hugo Garcia-Garcia, Luis J. Galán-Wong, Benito Pereyra-Alférez, Luis C. Damas-Buenrostro, Esmeralda Pérez, and Juan Carlos Cabada. 2017. DOI: https://doi.org/10.1094/ASBCJ-2017-4294-01.]</ref>. | ||
| − | + | Species of yeast and bacteria that are considered beer spoilers include [[Brettanomyces|''Brettanomyces'']] species, numerous [[Lactobacillus|''Lactobacillus'']] species, ''Pediococcus damnosus'', ''Pectinatus'' species (anaerobe responsible for 20-30% of bacterial contaminations that produces acetic acid, [https://en.wikipedia.org/wiki/Propionic_acid propionic acid], acetoin, and 'rotten egg' like odors in contaminated beer), ''Megasphaera cerevisiae'' (7% of bacterial contaminations; inhibited below pH 4.1 and 2.8% ABV but can produce considerable amounts of [[Butyric Acid|butyric acid]] along with smaller amounts of acetic acid, caproic acid, [[Isovaleric Acid|isovaleric acid]], acetoin, and hydrogen sulphide), ''Selenomonas lactifex'', ''Zymophilus'' spp., [[Saccharomyces#Diastatic_strains_of_Saccharomyces_cerevisiae|diastatic strains of ''Saccharomyces cerevisiae'']], and some species from the ''Candida'' and ''Pichia'' genera. Most wild yeasts that can grow in beer in lab conditions are not considered largely impactful because of their limitatons to growing in the presence of ethanol or lack of oxygen, but they can become impactful on barrel aged beers where oxygen is present (''Candida'' species, ''Pichia'' species, ''Torulaspora delbrueckii'', ''Issatchenkia orientalis'', ''Kluyveromyces marxianus'', ''Debaryomyces hansenii'', ''Zygosaccharomyces bailii'', ''Zygosaccharomyces bisporus'', ''Schizosaccharomyces pombe'', and ''Kloeckera apiculata'') <ref name="Bokulich_2013">[http://mmbr.asm.org/content/77/2/157.full The Microbiology of Malting and Brewing. Nicholas A. Bokulich and Charles W. Bamforth. 2013. DOI: 10.1128/MMBR.00060-12]</ref>. Hop tolerant lactic acid bacteria make up the majority of contamination issues in breweries, with ''L. brevis'' making up more than half of the reported contaminations, and all lactic acid bacteria making up 60-90% of reported contaminations. A new species of ''Lactobacillus'' was recently identified called ''L. acetotolerans'' and was [https://www.facebook.com/groups/MilkTheFunk/permalink/1363048380390039/ responsible for contaminating Goose Island's Bourbon County Stout], which is 60 IBU and 11% ABV. In sour beers with a pH below 4.3, only some lactic acid bacteria, ''Brettanomyces'', and some wild ''Saccharomyces cerevisiae'' strains (which sometimes produce phenols, haze, over-attenuation, and/or over-carbonation) have the potential for unwanted growth, while beers with low alcohol, a small amount of hops, lower CO<sup>2</sup> volumes (cask ales and beers dispensed with nitrogen, for example), and higher pH (4.4-4.6) are the most susceptible to contamination <ref name="Vaughan_2005">[https://onlinelibrary.wiley.com/doi/full/10.1002/j.2050-0416.2005.tb00221.x Enhancing the Microbiological Stability of Malt and Beer — A Review. Anne Vaughan, Tadhg O'Sullivan, Douwe Van Sinderen. 2005. DOI: https://doi.org/10.1002/j.2050-0416.2005.tb00221.x.]</ref>. ''Pectinatus'' and ''Megasphaera'' are Gram-negative anaerobic species that produce a number of off-flavors in ales but not lagers (probably due to their preference for warmer temperatures). They are somewhat tolerant of hops (they can grow in beers with IBU's as high as 33-38 with one strain isolated from pickles reported to grow in beer up to 5% ABV and 80 IBU <ref>[https://www.sciencedirect.com/science/article/pii/S0740002020300514 Comparative genetic and physiological characterisation of Pectinatus species reveals shared tolerance to beer-associated stressors but halotolerance specific to pickle-associated strains. Timo Kramer, Philip Kelleher, Julia van der Meer, Tadhg O’Sullivan, Jan-Maarten A.Geertman, Sylvia H. Duncan, Harry J. Flint, Petra Louis. 2020. DOI: https://doi.org/10.1016/j.fm.2020.103462.]</ref>) and often survive within the biofilms of other species in the brewing environment where the biofilm creates an anaerobic environment for them. They are sometimes found contaminating low ABV beers (under 5.2%) during packaging. They are not tolerant of pH below 4 and are killed at relatively low temperatures (58–60°C for one min) <ref name="Suzuki_2012" />. ''Zymomonas mobilis'' is a microaerophilic Gram-negative acetic acid bacteria that can withstand hops and can grow in bottled beer or casks where priming sugar is added and small amounts of air is present and produces high levels of acetaldehyde and hydrogen sulphide <ref name="Vaughan_2005" />. While the Gram-positive ''Staphylococcus xylosus'' bacteria, which grows on the skin of humans and animals, is not normally considered a beer contaminant, one strain was isolated from craft beer and was identified as the cause of increased turbidity, lactic acid, and succinic acid. It could grow at a pH between 3-7 (although only produced turbidity at a pH of 4-7), a temperature between 4–37°C, and as much as 8% ABV. This demonstrates that it is possible for species to adapt to living in beer other than the more typical beer spoilers <ref>[https://onlinelibrary.wiley.com/doi/pdf/10.1002/fsn3.1256 Beer‐spoilage characteristics of Staphylococcus xylosus newly isolated from craft beer and its potential to influence beer quality. Zhimin Yu, Qiuying Luo, Li Xiao, Yumei Sun, Rong Li, Zhen Sun, Xianzhen Li. 2019. DOI: 10.1002/fsn3.1256.]</ref>. | |
| − | |||
| − | |||
| − | + | Other species of microbes do not grow in beer but can become contaminants earlier on in the brewing process prior to primary fermentation (for example during kettle souring). These species include enterobacteria such as species of ''Clostridium'', ''Obesumbacterium'', ''Citrobacter'', and ''Rahnella aquatilis'', and wild ''Saccharomyces'' species that might not be able to grow in finished beer. These microbes produce [[Dimethyl Sulfide|dimethyl sulfide]], organic acids, and 2,3-butanediol in high amounts which gives the beer an unpleasant rotting fruity or vegetal aroma <ref name="Bokulich_2013" />. Other species are considered "indicator" species because they do not directly cause spoilage of beer, but indicate that there is a hygiene problem. These include ''Acetobacter'', ''Gluconobacter'', ''Escherichia '', and ''Klebsiella'', as well as aerobic yeasts, all of which usually don't have an impact when present unless oxygen is also present. They can also produce slime that protects other microorganisms that can have a greater impact on the beer's stability <ref name="Wirtanen_2001">[https://www.researchgate.net/publication/273439407_Disinfectant_testing_against_brewery-related_biofilms. Disinfectant testing against brewery-related biofilms. Erna Storgårds, Gun Wirtanen. 2001.]</ref><ref name="Bokulich_2018">[https://www.tandfonline.com/doi/abs/10.1094/ASBCJ-2012-0709-01 A Review of Molecular Methods for Microbial Community Profiling of Beer and Wine. Nicholas A. Bokulich, Charles W. Bamforth & David A. Mills. 2018.]</ref>. Some species can contaminate yeast pitches. ''Pediococcus damnosus'' is frequently the cause of such contaminations and can cause diacetyl problems, as well as ''Pediococcus inopinatus'', ''Pediococcus claussenii'', ''L. casei'', ''Selenomonas lacticifex'', and ''Zymophilus raffinosivorans'' (although these are rarer to find in finished beer). ''Obesumbacterium proteus'' (which gives a parsnip-like smell and flavor) and ''Rahnella aquatilis'' can contaminate yeast pitches, and can inhibit fermentation and result in the beer finishing at a higher pH <ref name="Vaughan_2005" />. | |
| − | + | Biofilm forming spoilage organisms include a much wider range and higher frequency in beer tap systems than in brewhouses. This is due to the availability of oxygen and higher temperatures at certain points in the tap system, as well as poorer hygiene in tap systems as well as the difficulty to effectively clean plastic hoses. Of particular concern here is the ability of ''E. coli'' serotype O157:H7 to survive in tap systems, which has had a couple of documented occurrences in contaminated apple cider. Another study showed that aerobic yeasts were able to grow in dispensing lines, as well as ''L. brevis'', and in many cases the draft lines were re-contaminated one week after cleaning, indicating that a contamination in draft lines is difficult to remove <ref name="storgards_2000">[http://www.vtt.fi/inf/pdf/publications/2000/P410.pdf Process hygiene control in beer production and dispensing. Erna Storgårds. VTT Publications 410. 2000.]</ref><ref name="Wirtanen_2001" />. | |
| − | + | Sources for contamination in breweries can occur as "primary" contaminations (yeast pitching, and brewhouse related contaminations), or as "secondary" contaminations (packaging and cellaring), as well as in tap systems. They are usually not sudden occurrences, but a result of the continued growth of microorganisms in a problem area. Historically, re-pitching yeast was often a source of contamination; however, more recently this has become less of a source for contaminations due to better education and techniques. Typical sources for contamination also include unclean equipment such as thermometers, manometers, valves, dead ends, gas pipes, leaks in any part of the system (especially at heat exchangers), wort aeration equipment, and even worn floor surfaces. More than half of the documented contaminations come from the packaging system. These are typically the sealer (35%), the filler (25%), the bottle inspector (10%), dripping water from the bottle washer (10%), and the environment close to the filler and sealer (10%). In regards to the environment as a source of contamination, this has been found to be from airborne contaminants near the filler and crowner in open bottles on their way from the bottle washer to the filler and from the filler to the capper. The higher the humidity and the more airflow, the more chances of airborne contamination <ref name="storgards_2000" /><ref name="Vaughan_2005" />. For example, ''Pectinatus'', while mostly found in lubrication oils, water systems, floors, water condensed on ceilings, etc., it can also survive on [https://en.wikipedia.org/wiki/Aerosol aerosols] in the air and is thought to possibly transferred to beer that is being packaged via the air <ref name="Suzuki_2012" />. In tap systems at taverns, 'one-way' valves that are attached to kegs have been found to be a source of contamination, as well as the dispensing line <ref name="storgards_2000" /><ref name="Vaughan_2005" />. | |
| − | |||
| − | Bacteria and yeast form a biofilm in two stages, which are determined by a number of variables. In the first stage, the microbes remain in their [http://www.dictionary.com/browse/planktonic|"planktonic"] form (floating around in the liquid), but they begin to adhere on surfaces and to each other as those surfaces. Other species of microbes can also be adhered to during this phase. The second stage is where the microbes start producing exopolysaccharides (EPS) which helps them bind together in a matrix, along with any available proteins and exopolymers produced by the bacteria. A large portion of biofilms is actually water (80-80%) as this allows the microbes to remove waste and consume nutrients. This matrix helps the microbes resist antibiotics, UV radiation, and cleaning chemicals. Gene exchange also occurs more frequently. At the end of this second stage, the microbes become attached to surfaces in such a way that is permanent without the use of cleaning chemicals. This is known as the microbe's [http://www.dictionary.com/browse/sessile|"sessile"] form (immobile). Bacteria in this form continue to multiply, and upon maturation of the biofilm, eventually, planktonic cells begin to be produced and released from the biofilm to find new homes. They also display different phenotypes, which might contribute to their ability to resist cleaning chemicals. Rough surfaces, scratched surfaces, jagged edges, and pores are more prone to biofilm formation due to the higher surface area. Hydrophobic surfaces, such as Teflon and other plastics, are more prone to biofilm formation than hydrophilic surfaces (glass and stainless steel). Nitrile butyl rubber (NBR) was found to inhibit biofilm formation when new, but as the material breaks down biofilms are able to grow <ref>Biofilms in the Food and Beverage Industries. P M Fratamico, B A Annous, N W Guenther. Elsevier, Sep 22, 2009. Pp 4-14.</ref>. Biofilm formation is strain specific rather than species specific; some strains can form thicker biofilms than others within the same species and faster, and some strains of lactic acid species are not good biofilm producers. Full biofilms can form within 2-4 days for some strains, while 10 days is required for significant biofilm formation in other strains. For example, one strain of ''Lactobacillus brevis'' isolated from draft beer did not form any biofilm, while another strain of ''L. brevis'' tested was a strong biofilm producer. Similar results were observed for ''Brettanomyces'' strains. In general, mixed cultures form stronger biofilms than single cultures. The presence of soil (biological residue) encourages biofilm formation <ref name="Wirtanen_2001" />. The presence of sweeteners or sugar also encourages the formation of biofilms. In one study (Storgårds 2006), biofilm forming species were found to begin attaching themselves to brand new sterile stainless steel surfaces within 2-12 hours after the new equipment was used for production <ref name="Storgårds_2006">[https://www.researchgate.net/publication/279707988_Microbial_attachment_and_biofilm_formation_in_brewery_bottling_plants Microbial attachment and biofilm formation in brewery bottling plants. Erna Storgårds, Kaisa Tapani, Peter Hartwall, Riitta Saleva & Maija-Liisa Suihko. 2006. DOI: https://doi.org/10.1094/ASBCJ-64-0008.]</ref>. | + | Thompson et al. (2024) divided the brewing environment into three "zones": areas that have direct contact with beer, such as the brewhouse, areas that are adjacent to beer production areas that have no physical barrier from the beer production area, and areas that have a physical barrier between the beer production areas, such as offices, tap rooms, etc. They took samples from all areas in three small microbreweries. The study found that racking arm valves were one of the worst offenders of housing contaminating microbes, although the results were skewed by one of the breweries having a higher load of contaminants in general between the three microbreweries. Proper cleaning, sanitation, and maintenance of racking arms is recommended to reduce biofilm formation. Other areas that harbored bacteria included the bottom valve, carb stone valve, spray ball valve, and the yeast pitch. All areas of the brewery that were swabbed show at least some bacterial growth, demonstrating that all areas of the brewery are potential sources of contamination. This study noticed that one brewery in particular had a higher occurrence of spoilage microbes; the study speculated that this might be due to cleaning and sanitation protocols during packaging, and heavy loads of contaminants were found in the packaging system <ref>[https://academicjournals.org/journal/JBD/article-full-text-pdf/705951B71856 Alex R. Thompson, Julie K. Northcutt and Paul Dawson. Bacterial contamination and surface hygiene in the microbrewery environment. Journal of Brewing and Distilling. Vol. 13(1), pp. 1-10, January-June 2024. DOI: 10.5897/JBD2024.0060.]</ref>. |
| + | |||
| + | See also: | ||
| + | * The [[Barrel#Sanitizing|Barrel]] wiki page. | ||
| + | * [http://mmbr.asm.org/content/77/2/157/F3.expansion.html Diagram overview of bacterial and fungal species reported at all stages of beer production.] | ||
| + | |||
| + | ==Microbe Populations in Mixed Fermentation Breweries== | ||
| + | |||
| + | Bokulich et al. (2015) studied the microbial populations throughout a brewery (presumably Allagash) that produces clean beer, mixed fermentation sour beer, and spontaneously fermented coolship ale. They found that most of the microbes living in breweries were introduced from the ingredients such as malted barley and hops, and many populations were confined to specific rooms or areas within the brewery. Some species did spread to other rooms, presumably through human and insect vectors. Beer resistant lactic acid bacteria spread throughout the brewery (although more abundantly found near packaging equipment and fermenters that were filled with sour beer), but the clean beer was largely uncontaminated. Physical partitions and walls appeared to help inhibit the spread of microbes from room to room <ref name="Bokulich_2015">[https://elifesciences.org/articles/04634 Mapping microbial ecosystems and spoilage-gene flow in breweries highlights patterns of contamination and resistance. Nicholas A Bokulich, Jordyn Bergsveinson, Barry Ziola, David A Mills. 2015.]</ref>. | ||
| + | |||
| + | Ale yeast was found throughout the brewery, especially in the fermentation cellar. Malted grains were determined to be the highest source of potentially contaminating microbes (mostly ''Pediococcus'', although the PCR method used in the study was not adequate for detecting microbes on grain) in the hotside areas of the brewery compared to other microbe sources such as human skin, outdoor air, soil, saliva, feces, water from the plumbing, etc <ref name="Bokulich_2015" />. Other studies have identified grain dust and aerosols as airborne vectors that disperse a range of lactic acid bacteria <ref name="Bokulich_2013" />. Hops were determined to be the highest source of microbes in the cellar fermentation areas (''Pediococcus'' spp, ''Lactobacillus lindneri'', and ''L. brevis'' were detected on pellet hops, although the presence of hop tolerant genes was not detected from microbes found on hops), and yeast was the highest population in fermenter and packaging areas. While human skin was a minor contributor to microbe populations found in the brewery, oak barrel surfaces were populated with microbes from unknown sources. Therefore, it was determined in this study that raw materials are the major source for potential contaminants in a brewery, although minor sources such as sinks are still potential problems and hop tolerant species were linked to the purposeful inoculation of souring microbes in the sour beer production itself <ref name="Bokulich_2015" />. ''L. lindneri'' is highly hop tolerant, difficult to detect on MRS media, and has been found to survive suboptimal heat pasteurization temperatures. ''L. brevis'' strains that have been growing in beer are also hop tolerant, although some strains have been found to lose their ability to grow in beer if they are grown in lab media after a few generations and strains not found in breweries are not hop tolerant, which suggests that strains found in beer have adapted to that environment. Both of these species also have a smaller cell size when they have been growing in beer, and can pass through microbial filtration systems <ref name="Suzuki_2012" />. | ||
| + | |||
| + | Seasonality played a minor role in the populations of microbes throughout the brewery, with ale yeast and ''Candida santamariae'' spreading from the fermentation and packaging area to the rest of the brewery from fall to summer (it was proposed that the warming from fall to summer played a role in the spread of ale yeast and ''Candida santamariae'' throughout the brewery). ''Micrococcus'' and ''Kocuria'' were found in more localized areas such as the floors and other surfaces in the barrel room, cellar, and packaging room. ''Acetobacter'' and ''Lactobacillus'' were found specifically in areas where a lot of wort or beer was being processed (conveyor belts and floors below the packaging equipment, hotside and cellar area sinks, and sample ports on kegs and fermenters). ''Lactobacillus'' was more common on surfaces where sour beer production was (fermenters and barrel surfaces), with floor surfaces having a more diverse mixture of LAB species <ref name="Bokulich_2015" />. | ||
| + | |||
| + | Microbes with hop tolerant genes were found more abundantly in the fermenter and packaging areas (filler heads, below the bottling line belt, packaging sink, and a keg faucet) compared to microbes found on pellet hops (which were determined to not be a source of contamination), kegs, or barrel bungs, and was associated with where beer was being processed, particularly mixed fermentation and spontaneously fermented sour beer <ref name="Bokulich_2015" />. | ||
| + | |||
| + | In the case of mixed cultures or contaminations that contain ''Pediococcus damnosus''., ''Lactobacillus brevis'', or ''Lactobacillus lindneri'', and possibly other species of these two genera, it is possible for the cells of these bacteria to adhere to the cells of brewers yeast. This can cause the yeast to prematurely drop out of suspension during fermentation, resulting in under attenuated beer. This function of the bacteria is thought to contribute to the slower development of the bacteria after the yeast has dropped out of solution. Other species of ''Lactobacillus'' such as ''L. casei/paracasei'', ''L. coryniformis'' and ''L. plantarum'' as well as species of ''Leuconostoc'' are very intolerant of hops and are therefore only considered a threat against beer that is very lowly hopped <ref name="Suzuki_2012" />. | ||
| + | |||
| + | ==Biofilms== | ||
| + | Many microorganisms can form ''biofilms'' which is defined as a community of cells of one or more species that are attached to each other and/or a surface and are embedded in a matrix of extracellular polymeric substances (EPS). EPS consists of polysaccharides and proteins that are produced by the microorganisms and expelled out of the cells, similar to a [[Pellicle|pellicle]]. Biofilms allow microbes to survive less vigorous cleaning and sanitizing regiments and chemicals and has become a concern in the food industry as well as in the brewing and winemaking industries <ref>[https://onlinelibrary.wiley.com/doi/abs/10.1111/1541-4337.12087 The Paradox of Mixed‐Species Biofilms in the Context of Food Safety. Iqbal Kabir Jahid and Sang‐Do Ha. 2014.]</ref>. Biofilms most often form in the packaging system somewhere, but can also be found on side rails, wearstrips, conveyor tracks, drip pans, and in-between chain links <ref name="storgards_2000" />. | ||
| + | |||
| + | Bacteria and wild yeast form a biofilm in two stages, which are determined by a number of variables. In the first stage, the microbes remain in their [http://www.dictionary.com/browse/planktonic|"planktonic"] form (floating around in the liquid), but they begin to adhere on surfaces and to each other as those surfaces. Other species of microbes can also be adhered to during this phase. The second stage is where the microbes start producing exopolysaccharides (EPS) which helps them bind together in a matrix, along with any available proteins and exopolymers produced by the bacteria. A large portion of biofilms is actually water (80-80%) as this allows the microbes to remove waste and consume nutrients. This matrix helps the microbes resist antibiotics, UV radiation, and cleaning chemicals. Gene exchange also occurs more frequently. At the end of this second stage, the microbes become attached to surfaces in such a way that is permanent without the use of cleaning chemicals. This is known as the microbe's [http://www.dictionary.com/browse/sessile|"sessile"] form (immobile). Bacteria in this form continue to multiply, and upon maturation of the biofilm, eventually, planktonic cells begin to be produced and released from the biofilm to find new homes. They also display different phenotypes, which might contribute to their ability to resist cleaning chemicals. Rough surfaces, scratched surfaces, jagged edges, and pores are more prone to biofilm formation due to the higher surface area. Hydrophobic surfaces, such as Teflon and other plastics, are more prone to biofilm formation than hydrophilic surfaces (glass and stainless steel). Nitrile butyl rubber (NBR) was found to inhibit biofilm formation when new, but as the material breaks down biofilms are able to grow <ref>Biofilms in the Food and Beverage Industries. P M Fratamico, B A Annous, N W Guenther. Elsevier, Sep 22, 2009. Pp 4-14.</ref>. Biofilm formation is strain specific rather than species specific; some strains can form thicker biofilms than others within the same species and faster, and some strains of lactic acid species are not good biofilm producers <ref name="Wirtanen_2001" />. Wild strains of ''S. cerevisiae'' that carry the "Flo11p" gene tend to form biofilms which suggests that this ability is important for survival in the wild, but domesticated strains have mostly lost this ability probably due to evolving under nutrient-rich environments (human-controlled fermentation), and their planktonic form may give them an advantage in nutrient-rich liquids, especially during spontaneous fermentation where their ability to be mobile might help them compete against other species of microbes <ref>[https://www.nature.com/articles/s41467-018-05106-7 The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Shou-Fu Duan, Pei-Jie Han, Qi-Ming Wang, Wan-Qiu Liu, Jun-Yan Shi, Kuan Li, Xiao-Ling Zhang & Feng-Yan Bai. 2018.]</ref><ref>[https://www.ncbi.nlm.nih.gov/pubmed/11157168 Bakers' yeast, a model for fungal biofilm formation. Reynolds and Fink. 2011. DOI: 10.1126/science.291.5505.878.]</ref>. Full biofilms can form within 2-4 days for some strains, while 10 days is required for significant biofilm formation in other strains. For example, one strain of ''Lactobacillus brevis'' isolated from draft beer did not form any biofilm, while another strain of ''L. brevis'' tested was a strong biofilm producer. Similar results were observed for ''Brettanomyces'' strains. In general, mixed cultures form stronger biofilms than single cultures. The presence of soil (biological residue) encourages biofilm formation <ref name="Wirtanen_2001" />. The presence of sweeteners or sugar also encourages the formation of biofilms. In one study (Storgårds 2006), biofilm forming species were found to begin attaching themselves to brand new sterile stainless steel surfaces within 2-12 hours after the new equipment was used for production <ref name="Storgårds_2006">[https://www.researchgate.net/publication/279707988_Microbial_attachment_and_biofilm_formation_in_brewery_bottling_plants Microbial attachment and biofilm formation in brewery bottling plants. Erna Storgårds, Kaisa Tapani, Peter Hartwall, Riitta Saleva & Maija-Liisa Suihko. 2006. DOI: https://doi.org/10.1094/ASBCJ-64-0008.]</ref>. | ||
The efficacy of different chemicals to kill microbes within a biofilm isn't widely studied in the brewing or wine industries, partly because testing procedures are laborious and difficult to standardize. Studies have found that alcohol-based disinfectants (ethanol and isopropyl alcohol) and hydrogen peroxide-based disinfectants were effective at killing microbes within a biofilm, and peracetic acid disinfectants were not as effective. A higher concentration of peracetic acid (from 0.25% to 1% of products containing 4-15%) was required to be more effective than lower concentrations. However, these disinfectants did not kill all of the cells without a cleaning regiment first. Yeast biofilms, in general, are more susceptible to cleaning chemicals than bacteria biofilms. Biofilms that are formed under static conditions (still or dried up liquid) are more resistant to disinfectants than biofilms that form under flow conditions (movement of liquid) <ref name="Wirtanen_2001" /><ref name="Wirtanen_2003">[https://link.springer.com/article/10.1023/B:RESB.0000040471.15700.03 Disinfection in Food Processing – Efficacy Testing of Disinfectants. G. Wirtanen, S. Salo. 2003.]</ref>. | The efficacy of different chemicals to kill microbes within a biofilm isn't widely studied in the brewing or wine industries, partly because testing procedures are laborious and difficult to standardize. Studies have found that alcohol-based disinfectants (ethanol and isopropyl alcohol) and hydrogen peroxide-based disinfectants were effective at killing microbes within a biofilm, and peracetic acid disinfectants were not as effective. A higher concentration of peracetic acid (from 0.25% to 1% of products containing 4-15%) was required to be more effective than lower concentrations. However, these disinfectants did not kill all of the cells without a cleaning regiment first. Yeast biofilms, in general, are more susceptible to cleaning chemicals than bacteria biofilms. Biofilms that are formed under static conditions (still or dried up liquid) are more resistant to disinfectants than biofilms that form under flow conditions (movement of liquid) <ref name="Wirtanen_2001" /><ref name="Wirtanen_2003">[https://link.springer.com/article/10.1023/B:RESB.0000040471.15700.03 Disinfection in Food Processing – Efficacy Testing of Disinfectants. G. Wirtanen, S. Salo. 2003.]</ref>. | ||
See also: | See also: | ||
| + | * [[Brettanomyces#Biofilm|''Brettanomyces'' biofilm]] | ||
* [https://twitter.com/socialmicrobes/status/983764240254341125?s=04 Time lapse of biofilm formation.] | * [https://twitter.com/socialmicrobes/status/983764240254341125?s=04 Time lapse of biofilm formation.] | ||
| − | + | ==Spores== | |
| − | Some species of fungi and bacteria can form spores. | + | Some species of fungi and bacteria can form spores. Yeast generally forms spores in order to reproduce sexually. Their sporulated forms are not a mode of protection from disinfectants and are therefore killed by normal sanitation methods. Bacteria and molds form spores as a mode of survival. For example, some dangerous types ''Clostridium botulinum'' spores require 250°F (121°C) for 3 minutes to be killed, which is the requirement for canned goods <ref>[https://www.cdc.gov/mold/faqs.htm#mold "What are molds?". CDC website. Retrieved 01/25/2019.]</ref><ref>[http://www.jfoodprotection.org/doi/abs/10.4315/0362-028X-45.5.466?code=fopr-site Differences and Similarities Among Proteolytic and Nonproteolytic Strains of Clostridium botulinum Types A, B, E and F: A Review. RICHARD K. LYNT*, DONALD A. KAUTTER and HAIM M. SOLOMON. 1982.]</ref><ref>[http://beerandwinejournal.com/botulism/ Chris Colby. "Storing Wort Runs the Risk of Botulism". Beer and Wine Journal Blog. 04/17/2014. Retrieved 04/04/2018.]</ref>. Spore-forming species of bacteria, however, are not considered beer spoilers <ref>[https://www.facebook.com/groups/MilkTheFunk/permalink/2047716495256554/?comment_id=2047776558583881&reply_comment_id=2048688798492657&comment_tracking=%7B%22tn%22%3A%22R%22%7D Bryan Heit. Milk The Funk Facebook thread on yeast and bacteria spores and brewery hygiene. 04/04/2018.]</ref>. Thus, the challenge of killing yeast or bacteria spores is irrelevant in most beer and wine production. There are some extraneous brewing methods where bacteria spores should be considered, for example [[Wild_Yeast_Isolation#Safety|wild yeast isolation safety]], [[mold]] formation during fruit fermentation or barrel aging, and the [http://beerandwinejournal.com/botulism/ long storage of unfermented wort]. ''Lactobacillus'' species [https://www.researchgate.net/post/How_to_prepare_spore_forming_media_for_lactobacillus do not form spores]. |
| − | == | + | ==Hygienic Methods For Avoiding Contamination== |
| − | + | ===General Approaches=== | |
| − | * Use 180°F | + | * Clean and disinfect all equipment that comes into contact with the beer or wort, including beer dispensing systems, and keep them in prime condition. |
| − | * For any plastics that cannot be treated with heat, especially tubing, keep separate plastics for use with ''Lactobacillus'', ''Brettanomyces'', and ''Saccharomyces | + | * Use the maximum concentrations, exposure times, and hottest temperatures (considering temperature limitations of plastics and glass) suggested by the manufacturers of any given cleaning/disinfectant product. |
| − | * Replace rubber and plastic parts such as gaskets as often as recommended by the manufacturer. | + | * Clean first using an effective cleaner, and then apply a disinfectant/sanitizer as a second step. |
| − | * When operating a commercial brewery | + | * Use 180°F (82°C) hot water for 60 minutes to disinfect stainless steel and other heat tolerant materials (check with your manufacturer to make sure that the vessel is rated to withstand fast hot/cold cycles; vacuum or pressure relief valves should be used in order to avoid imploding due to fast temperature shifts for some equipment). |
| − | * Use a separate packaging system for sour beer unless the packaging system can be sanitized with hot water or caustic (foam disinfectants that are often used in packaging lines have been reported to be not as effective against removing biofilms). | + | * For any plastics that cannot be treated with heat, especially tubing, keep separate plastics for use with potential contaminants such as ''[[Lactobacillus]]'', ''[[Pediococcus]]'', ''[[Brettanomyces]]'', and ''[[Saccharomyces#Diastatic_strains_of_Saccharomyces_cerevisiae|diastatic strains of ''Saccharomyces cerevisiae'']]''. |
| + | * Replace rubber and plastic parts such as gaskets as often as recommended by the manufacturer or when wear is apparent. | ||
| + | * When operating a commercial brewery, invest in a quality control lab and procedures to identify inefficient hygiene practices. | ||
| + | * Use a separate packaging system for sour beer unless the packaging system can be sanitized with hot water or caustic (foam disinfectants that are often used in packaging lines have been reported to be not as effective against removing biofilms). | ||
| + | * The more surface area that equipment has, the more prone it is to biofilm formation. Horizontal surfaces are more prone than vertical surfaces to biofilm formation. | ||
| + | * Heat pasteurize, and store beer and yeast at low temperatures. Beer filtration and pasteurization are effective ways to reduce the chance of contamination. | ||
| + | * Non-alcoholic beer and beer under 1.3% ABV grew bacteria 2-5 times more than 4.5% ABV lager beer in one study, and the authors concluded that long-draw draft lines should not be used to serve these types of beers <ref>[https://onlinelibrary.wiley.com/doi/abs/10.1002/jib.670 Quain, D. E. (2021) The enhanced susceptibility of alcohol-free and low alcohol beers to microbiological spoilage: implications for draught dispense, J. Inst. Brew., XXX, doi: https://doi.org/10.1002/jib.670.]</ref>. | ||
| − | + | ===Reducing Microorganisms=== | |
| − | Several generalized procedures are used for limiting the number of unwanted microorganisms. These include acid washing yeast that is re-pitched (kills bacteria but not wild yeast), keeping beer cool (slows the growth of microbes in general), filtration (removes yeast), pasteurization (kills vegetative cells in the finished beer, but not spores - most beer spoilers are killed at 15 [http://wiki.zero-emissions.at/index.php?title=Pasteurization_in_beer_production pasteurization units (PU)] and all are killed at 30 PU using a recommended pasteurization temperature of 66°C ), and aseptic or hygienic packaging. Packaging systems should be frequently flooded with hot water between 80-95°C or saturated steam (every 2 hours in the summer and every 4 hours in the winter). UV light or disinfecting chemicals are also used. The filler and crowner should be disinfected frequently as well. Packaging in an aseptic room with HEPA filtration and higher air pressure within the room compared to outside, along with special clothing, is another method that larger breweries use to remain aseptic <ref name="storgards_2000" />. | + | Several generalized procedures are used for limiting the number of unwanted microorganisms. These include acid washing yeast that is re-pitched (kills bacteria but not wild yeast that is tolerant of low pH), keeping beer cool (slows the growth of microbes in general), filtration (removes yeast), pasteurization (kills vegetative cells in the finished beer, but not spores - most beer spoilers are killed at 15 [http://wiki.zero-emissions.at/index.php?title=Pasteurization_in_beer_production pasteurization units (PU)] and all are killed at 30 PU using a recommended pasteurization temperature of 66°C ), and aseptic or hygienic packaging. Note that some strains of ''Lactobacillus'' have been shown to survive pasteurization temperatures; see [[Lactobacillus#Tolerance_of_Extreme_Temperature|''Lactobacillus'' Heat Tolerance]]. Packaging systems should be frequently flooded with hot water between 80-95°C or saturated steam (every 2 hours in the summer and every 4 hours in the winter). UV light or disinfecting chemicals are also used. The filler and crowner should be disinfected frequently as well. Packaging in an aseptic room with HEPA filtration and higher air pressure within the room compared to outside, along with special clothing, is another method that larger breweries use to remain aseptic <ref name="storgards_2000" />. |
| − | Most brewing equipment should be designed for good hygiene. Pits and crevices should be avoided, and all surfaces should be smooth when possible. All equipment and pipelines should be self-draining. Valves are a typical source of contamination because they are not easily CIP'ed, especially plug valves and ball valves (although butterfly, gate, and globe valves are also difficult to CIP) <ref name="storgards_2000" />. Horizontal surfaces and wet surfaces are more prone to biofilm formation. In one study that compared biofilm formation in bottling lines versus canning lines, it was found that canning lines develop less microbial biofilms and contaminations than bottling lines due to not having rinsing stations, labeling stations, and simpler constructions than the bottling lines that were studied <ref name="Storgårds_2006" />. However, some canning lines cannot use caustic for cleaning or it is not common practice but use foaming agents instead which are less effective at removing biofilms (see [[Quality_Assurance#Efficacy_of_Cleaning_Agents|efficacy of cleaning agents below]]). The lack of use of caustic cleaners in canning lines has been identified as a source of contamination issues with [[Saccharomyces# | + | Most brewing equipment should be designed for good hygiene. Pits and crevices should be avoided, and all surfaces should be smooth when possible. All equipment and pipelines should be self-draining. Valves are a typical source of contamination because they are not easily CIP'ed, especially plug valves and ball valves (although butterfly, gate, and globe valves are also difficult to CIP) <ref name="storgards_2000" />. Horizontal surfaces and wet surfaces are more prone to biofilm formation. In one study that compared biofilm formation in bottling lines versus canning lines, it was found that canning lines develop less microbial biofilms and contaminations than bottling lines due to not having rinsing stations, labeling stations, and simpler constructions than the bottling lines that were studied <ref name="Storgårds_2006" />. However, some canning lines cannot use caustic for cleaning, or it is not common practice, but use foaming agents instead which are less effective at removing biofilms (see [[Quality_Assurance#Efficacy_of_Cleaning_Agents|efficacy of cleaning agents below]]). The lack of use of caustic cleaners in canning lines has been identified as a source of contamination issues with [[Saccharomyces#Diastatic_strains_of_Saccharomyces_cerevisiae|diastatic strains of ''Saccharomyces cerevisiae'']] in canning lines <ref>[https://www.facebook.com/groups/MilkTheFunk/permalink/1561762887185253/?comment_id=1791471917547681&reply_comment_id=2017381731623364&comment_tracking=%7B%22tn%22%3A%22R9%22%7D Caroline Smith from Lallemand. Milk The Funk Facebook group post on diastaticus contamination. Feb 2018.]</ref>. |
| − | ====Cleaning and Sanitizing | + | Some other methods have been proposed by scientists as being novel ways to reduce unwanted microorganisms. These include exploiting naturally produced toxins. For example, some lactic acid bacteria [[Lactobacillus#Bacteriocins|produce bacteriocins]] which can kill other bacteria. Some strains of wine yeast can [[Saccharomyces#Killer_Wine_Yeast|produce zymocins]] that kill other species of yeast. Such methods are viewed as being fairly extreme. Advances in genetic engineering techniques make these approaches technically possible, however, there currently exists a commercial stigma against genetic modification. Additionally, there are many types of toxins which target only specific species, so anticipating which species should be targeted could be challenging <ref name="Vaughan_2005" />. |
| + | |||
| + | ====Yeast Washing==== | ||
| + | Yeast washing is the practice of exposing a yeast slurry to extreme acidic conditions in order to destroy bacteria contaminants in the slurry, and it has been used for over a century in the brewing industry to help reduce the potential for lactic acid bacteria spoilage. Although techniques might vary throughout the brewing industry, the most typical technique is to add phosphoric acid to a slurry of yeast until a pH of 2 is reached, and then the slurry is stored for 2 hours at 5°C (41°F). While phosphoric acid is a good choice for acid washing because of being inexpensive compared to other acids, its tendency to not kill yeast, and its lack of affecting beer flavor, it also does not kill some contaminants such as ''Shimwellia pseudoproteus''. It has also been proposed that chlorine dioxide, a disinfectant that is often used in the vegetable, meat, and water treatment industries, can be successfully used to wash a yeast slurry, with the first study on this reporting that a concentration of 78 mg/L (concentration value is for the entire slurry) and stored for 30 minutes at 8°C (46.4°F) was effective <ref>[https://link.springer.com/article/10.1007/s00253-020-10534-x Modeling the inactivation of Lactobacillus brevis DSM 6235 and retaining the viability of brewing pitching yeast submitted to acid and chlorine washing. Munford, A.R.G., Chaves, R.D., Granato, D. et al. Appl Microbiol Biotechnol (2020). https://doi.org/10.1007/s00253-020-10534-x.]</ref>. | ||
| + | |||
| + | Lysozyme, an enzyme that is often extracted from hen egg whites, is known to inhibit Gram-negative bacteria such as ''Lactobacillus'' but not Gram-positive bacteria such as ''Acetobacter'', and has been shown to be an enzyme that can help inhibit spoilage bacteria in wine and cider fermentations <ref>[https://en.wikipedia.org/wiki/Lysozyme "Lysozyme". Wikipedia. Retrieved 0319/2020.]</ref>. Lysozyme is normally added to wine with a stuck fermentation or to limit malolactic fermentation, and several yeast companies offer a lysozyme-based product <ref>[https://scottlab.com/content/files/Documents/Handbooks/ScottlabsHandbook2018.pdf 2018 Fermentation Handbook. Scott Laboratories. Retrieved 03/19/2020.]</ref><ref>[https://www.academia.edu/16244381/Lysozyme_in_Wine_An_Overview_of_Current_and_Future_Applications?email_work_card=title Lysozyme in Wine: An Overview of Current and Future Applications. Marco Esti, Ilaria Benucci. Comprehensive Reviews in Food Science and Food Safety. 2014.]</ref>. It has also been suggested to be useful for limiting lactic acid bacteria in yeast slurries, but one experiment reported that the sensitivity of different species of lactic acid bacteria varies, with ''Pediococcus inopinatus'', ''Lactobacillus brevis'', ''Lactobacillus brevisimilis'' showing similar levels of sensitivity, but ''L. linderi'' showing less sensitivity. Bacteria were inhibited more at 22°C than at 4°C. At 300 mg/L, although lactic acid bacteria was inhibited, it was not killed completely <ref>[https://www.researchgate.net/publication/293048096_Antibacterial_properties_of_hen_egg_white_lysozyme_against_beer_spoilage_bacteria_and_effect_of_lysozyme_on_yeast_fermentation/citation/download Van Landschoot, Anita & Villa, A. (2005). Antibacterial properties of hen egg white lysozyme against beer spoilage bacteria and effect of lysozyme on yeast fermentation.]</ref>. Nisin has also been proposed as a potential preservative that can be added to wort during boiling or cooling as well as to the fermenter in order to limit the growth of lactic acid bacteria by up to 90% <ref>[https://onlinelibrary.wiley.com/doi/full/10.1002/jib.233 Müller-Auffermann, K, Grijalva, F, Jacob, F, and Hutzler, M (2015), Nisin and its usage in breweries: a review and discussion. J. Inst. Brew., 121, 309–319. doi: 10.1002/jib.233.]</ref>. | ||
| + | |||
| + | The homebrew practice of mixing distilled or sanitized water into a yeast slurry, letting the slurry settle into three layers, and then removing the bottom and top layer and re-pitching or saving the middle layer, is different than "yeast washing". This process is known as "yeast rinsing", and is primarily employed by homebrewers who wish to separate trub material from their yeast slurries before reusing the yeast slurry. This might have the benefit of removing unwanted flavors from the slurry (although there is a lack of evidence that we know of for this claim <ref>[http://brulosophy.com/2015/03/02/sloppy-slurry-vs-clean-starter-exbeeriment-results/ "Sloppy Slurry vs. Clean Starter". Brulosophy website. 2015. Retrieved 03/19/2020.]</ref>) or hop material that could inhibit yeast growth, but it does not inhibit lactic acid bacteria or any other contaminants (in fact, this process increases the chances of contaminating the yeast slurry). See [https://www.homebrewersassociation.org/how-to-brew/yeast-washing-yeast-rinsing-whats-difference/ this AHA article] for more details on yeast rinsing. | ||
| + | |||
| + | ===Cleaning and Sanitizing=== | ||
The goal of cleaning is to remove as much biomaterial as possible, while the goal of sanitizing is to reduce the population of viable microbes as much as possible and prevent them from growing on surfaces during the non-production time. It's been shown that chemical cleaners are better at removing biofilms than sanitizers and disinfectants, and sanitizers that kill cells in suspension may not be effective at killing cells within biofilms. Complete removal of unwanted microbes within biofilms can be achieved by first using a cleaning agent to remove the biomass followed by a sanitizing/disinfecting agent. CIP procedures may not be enough to remove biofilms without high turbulent flow with spray nozzles and the use of heat (low cleaning temperatures are not effective at removing biofilms). Chlorinated alkaline detergents were found to be the most effective at removing biofilms <ref name="Wirtanen_2001" />. Below is a typical CIP process according to [http://www.vtt.fi/inf/pdf/publications/2000/P410.pdf Erna Storgårds (2000)]; CIP processes at room temperatures are not adequate enough to remove biofilms, so use hot temperatures when applicable. Use the highest chemical concentrations recommended by the vendor. Also, the higher the velocity of the cleaning fluid through the system, the more efficient it is at removing biofilms: | The goal of cleaning is to remove as much biomaterial as possible, while the goal of sanitizing is to reduce the population of viable microbes as much as possible and prevent them from growing on surfaces during the non-production time. It's been shown that chemical cleaners are better at removing biofilms than sanitizers and disinfectants, and sanitizers that kill cells in suspension may not be effective at killing cells within biofilms. Complete removal of unwanted microbes within biofilms can be achieved by first using a cleaning agent to remove the biomass followed by a sanitizing/disinfecting agent. CIP procedures may not be enough to remove biofilms without high turbulent flow with spray nozzles and the use of heat (low cleaning temperatures are not effective at removing biofilms). Chlorinated alkaline detergents were found to be the most effective at removing biofilms <ref name="Wirtanen_2001" />. Below is a typical CIP process according to [http://www.vtt.fi/inf/pdf/publications/2000/P410.pdf Erna Storgårds (2000)]; CIP processes at room temperatures are not adequate enough to remove biofilms, so use hot temperatures when applicable. Use the highest chemical concentrations recommended by the vendor. Also, the higher the velocity of the cleaning fluid through the system, the more efficient it is at removing biofilms: | ||
| Line 54: | Line 89: | ||
| Rinse || cold || 10-30 min | | Rinse || cold || 10-30 min | ||
|- | |- | ||
| − | | Disinfection (chemical such as peracetic acid or hot water at 85-90°C) || cold (or hot if using water) || 10-30 min with chemical, or 45-60 min with hot water | + | | Disinfection (chemical such as peracetic acid or hot water at 85-90°C) || cold (or hot if using just water) || 10-30 min with chemical, or 45-60 min with hot water |
|- | |- | ||
| Rinse (might contain a low concentration disinfectant) || cold || 5-10 min | | Rinse (might contain a low concentration disinfectant) || cold || 5-10 min | ||
| Line 61: | Line 96: | ||
Open surfaces such as bottle inspectors, fillers, and conveyor belts in the packaging line should be first rinsed with water, then cleaned with a foaming agent, rinsed again with water, and then sprayed with a disinfectant solution and a final rinse. Components that cannot be visually inspected should be dismantled and inspected. Rubber gaskets and sealings have been found to house biofilms, especially after deteriorating, and so they should be inspected and replaced as needed. NBR rubber has been found to inhibit biofilms when new, and EPDM rubber has been found to be anti-bacterial towards some bacteria <ref name="Wirtanen_2001" />. | Open surfaces such as bottle inspectors, fillers, and conveyor belts in the packaging line should be first rinsed with water, then cleaned with a foaming agent, rinsed again with water, and then sprayed with a disinfectant solution and a final rinse. Components that cannot be visually inspected should be dismantled and inspected. Rubber gaskets and sealings have been found to house biofilms, especially after deteriorating, and so they should be inspected and replaced as needed. NBR rubber has been found to inhibit biofilms when new, and EPDM rubber has been found to be anti-bacterial towards some bacteria <ref name="Wirtanen_2001" />. | ||
| − | See also: | + | See also: |
| + | * [https://www.masterbrewerspodcast.com/253 MBAA Podcast Episode 253 CIP Fundamentals.] | ||
| + | * [[Barrel#Sanitizing|Barrel Sanitizing]]. | ||
* [https://www.facebook.com/groups/MilkTheFunk/permalink/1891215887573283/ Joe Idoni's heat sanitation based SOP.] | * [https://www.facebook.com/groups/MilkTheFunk/permalink/1891215887573283/ Joe Idoni's heat sanitation based SOP.] | ||
* [https://www.facebook.com/groups/MilkTheFunk/permalink/1952066318154906/ MTF thread on brewing mixed fermentation beers with clean beers in a commercial brewery.] | * [https://www.facebook.com/groups/MilkTheFunk/permalink/1952066318154906/ MTF thread on brewing mixed fermentation beers with clean beers in a commercial brewery.] | ||
* [https://www.facebook.com/groups/MilkTheFunk/permalink/1710242802337260/ Another MTF thread on sanitation.] | * [https://www.facebook.com/groups/MilkTheFunk/permalink/1710242802337260/ Another MTF thread on sanitation.] | ||
| + | * [https://www.facebook.com/groups/MilkTheFunk/permalink/2461516447209888/ MTF thread on using one keg cleaner to clean regular beers and sour beers in commercial breweries.] | ||
| + | * [https://byo.com/article/a-clean-fight-the-science-of-hygienic-brewing/ "A Clean Fight: The Science of Hygienic Brewing" by Colin Kaminski in Brew Your Own Magazine.] | ||
| + | *[https://www.facebook.com/groups/MilkTheFunk/permalink/4279204685441046/ Brian Hall demonstrates steam sanitizing a corny keg.] | ||
| + | * [https://www.theseus.fi/bitstream/handle/10024/348696/Assessing%20Contamination%20Risk%20of%20Non-Conventional%20Yeast%20in%20Breweries.Ronja%20Eerik%C3%A4inen.pdf?sequence=2 "Assessing Contamination Risks of Non-Conventional Yeasts in Breweries," bachelors thesis for Ronja Eerikäinen (yeast study only)]; see also an interview with Eerikäinen on the [https://brulosophy.com/podcasts/the-bru-lab/ The Brü Lab podcast episode 16]. | ||
| + | * [https://www.birkocorp.com/wp-content/uploads/2017/08/Birko_ReducingDissolvedOxygen_WhitePaper.pdf "Reducing Dissolved Oxygen: Acid and Detergent Cleaning of Brite Tanks," white paper by Dana Johnson Technical Director, Craft Brewing, Birko. As appeared in The New Brewer, July/August 2011.] | ||
| + | |||
| + | ===Oak Barrels=== | ||
| + | |||
| + | See the [[Barrel#Sanitizing|Barrel]] wiki page. | ||
| + | |||
| + | ===Pasteurization=== | ||
| + | Pasteurization is measured in terms of "pasteurization units" (PU). One PU is equal to exposure of 60°C (140°F) for 1 minute. The total PU is determined by plotting time against temperature in degrees Fahrenheit. A total of 15 PU's has been given as the target for pasteurizing beer. The following equation can be used to calculate PU's using different temperatures and times <ref name="Haas_1960">[https://www.sciencedirect.com/science/article/abs/pii/S0065216408701242 Gerhard J. Haas. "Microbial Control Methods in the Brewery". Editor(s): Wayne W. Umbreit. Advances in Applied Microbiology. Academic Press, Volume 2, 1960. Pages 113-162. ISSN 0065-2164. ISBN 9780120026029.]</ref>: | ||
| + | |||
| + | [[File:PU calculation.jpg|none|360px|]] | ||
| + | |||
| + | There are two types of pasteurization methods used in brewing: tunnel pasteurization and flash pasteurization. In tunnel pasteurization, which is more widely used in breweries, cans or bottles of packaged beer is moved slowly through a tunnel of fixed temperatures. In flash pasteurization (or plate pasteurization), large quantities of beer are pasteurized at the same time via a heat exchanger and is usually performed before the beer is packaged <ref name="Vaughan_2005" />. Since thermal death rates for beer spoilage organisms has been identified to be 140°F (60°C) for 15 minutes <ref name="Haas_1960" /><ref>[https://onlinelibrary.wiley.com/doi/pdf/10.1002/j.2050-0416.1946.tb01593.x THERMAL DEATH POINTS OF MICRO-ORGANISMS IN BEER. Aage Lund. 1947.]</ref>, this is the baseline temperature and time for pasteurization, although higher temperatures and shorter times are used for some pasteurization methods (see the below links). The complete thermal death of ''Brettanomyces'' in wines has been reported to be 50°C for 5 minutes. <ref>[https://pubmed.ncbi.nlm.nih.gov/15996781/ Thermal inactivation of the wine spoilage yeasts Dekkera/Brettanomyces. José António Couto, Filipe Neves, Francisco Campos, Tim Hogg. 2005. DOI: 10.1016/j.ijfoodmicro.2005.03.014.]</ref><ref name="Nunes de Lima 2020" />. ''Pediococcus'' is generally not tolerant of temperatures over 45°C (see [[Pediococcus#Growth_and_Environment|''Pediococcus'']]). Some strains of ''Lactobacillus'' have been shown to potentially survive pasteurization temperatures for at least some amount of time; see [[Lactobacillus#Tolerance_of_Extreme_Temperature|''Lactobacillus'' Heat Tolerance]] for more information. | ||
| + | |||
| + | Microfiltration is an alternative technology to heat pasteurization that can be used to pasteurize beer. Microfiltration uses a set of membranes, usually in the 0.45–0.65 μm range, for filtering bacteria and yeast. Bacteria have a cell size of about 5-10 μm and yeast species have a cell size of about 5–16 μm, while flavor compounds such as phenols are filtered out when using a smaller diameter filter such as 0.2 μm. One study by Bernardi et al. (2019) found that filtration with polyethermide membranes removed around 1-2 IBU, ~30% of yeast-produced phenolic compounds (most polyphenols from hops were not filtered out), and larger tannins (which were only a small portion of the total polyphenol content). The antioxidant activity was largely not impacted. After filtration, the beers were 26%-33% lighter in color, depending on the style of the beer, and were 100% clearer. The filtration that was used, which was 1.2 μm, also produced fully pasteurized beers <ref>[https://www.sciencedirect.com/science/article/pii/B9780128152584000135 Microfiltration for Filtration and Pasteurization of Beers. Guilherme dos Santos Bernardi, Jacir Dal Magro, Marcio A. Mazutti, J. Vladimir Oliveira, Marco Di Luccio, Giovani Leone Zabot, Marcus V. Tres. 2019. DOI: https://doi.org/10.1016/B978-0-12-815258-4.00013-5.]</ref>. | ||
| + | |||
| + | Diastatic strains of ''Saccharomyces cerevisiae'' can have a wide range of temperature tolerance. Strains that can form ascospores or vegetative cells can be more heat tolerant. One thesis paper reported it taking 9 minutes at 60°C to kill 90% of a strain of diastatic ''S. cerevisiae'' in ascospore form <ref>[https://www.sciencedirect.com/science/article/pii/S2214799322000170#bib0135 Spoilage yeasts in beer and beer products. Inge M Suiker, Han AB Wösten. Current Opinion in Food Science. Microbiology, Department of Biology, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands. 02/19/2022.]</ref>. | ||
| + | |||
| + | See also: | ||
| + | * [[Barrel#Sanitizing|Barrel Sanitizing]]. | ||
| + | * [http://wiki.zero-emissions.at/index.php?title=Pasteurization_in_beer_production "Pasteurization in Beer Production"] and [http://wiki.zero-emissions.at/index.php?title=Process_info:_Pasteurization_in_beer_production "Pasteurization in Breweries"]; AEE - Institut für Nachhaltige Technologien wiki. | ||
| + | * [http://milkfacts.info/Milk%20Processing/Heat%20Treatments%20and%20Pasteurization.htm Heat Treatments and Pasteurization standards for milk processing.] | ||
| + | * [https://www.craftbrewingbusiness.com/packaging-distribution/preserve-product-quality-flash-pasteurization/ "Is flash pasteurization right for your craft beer?" by Chris Crowell in Craft Brewing Business website (details case studies for temperatures and times).] | ||
| + | * [https://www.masterbrewerspodcast.com/240 "Understanding the Risk of Can Pressure Failures" interview with Jim Kuhr on MBAA Podcast episode #240.] | ||
| + | * [https://www.masterbrewerspodcast.com/314 MBAA Podcast "Pasteurization At Goose Island".] | ||
| + | * [https://www.homebrewtalk.com/forum/threads/easy-stove-top-pasteurizing-with-pics.193295/?fbclid=IwAR3Glsqo-mWT70l4mY9AhmYa9SFKpfxo8gJAi8wJixlOlyccHVU5VCzn3cQ Example homebrew method for heat pasteurization by Pappers_ on HomebrewTalk '''(do not attempt this with highly carbonated beverages; bottles will break)'''.] | ||
| + | * [https://www.facebook.com/groups/MilkTheFunk/permalink/2350583941636473/ MTF thread on using sulfites and sorbate to stabilize fermentation in beer.] | ||
| + | * [[Cider#Keeving|"Keeving," a traditional method of achieving carbonation in sweet ciders (not pasteurization, but potentially a related application).]] | ||
| + | |||
| + | ===Draught Line Cleaning=== | ||
| + | See the Brewers Association [https://www.brewersassociation.org/educational-publications/draught-beer-quality-manual/ Draught Beer Quality Manual]. | ||
| − | + | ==Efficacy of Cleaning Agents== | |
| − | + | ===Commercial cleaners and disinfectants=== | |
[[File:Wirtanen disinfectants.PNG|thumb|300px|Advantages and disadvantages of disinfectants in food processing. Source: [https://link.springer.com/article/10.1023/B:RESB.0000040471.15700.03 "Disinfection in Food Processing – Efficacy Testing of Disinfectants". G. Wirtanen and S. Salo. 2003.]]] | [[File:Wirtanen disinfectants.PNG|thumb|300px|Advantages and disadvantages of disinfectants in food processing. Source: [https://link.springer.com/article/10.1023/B:RESB.0000040471.15700.03 "Disinfection in Food Processing – Efficacy Testing of Disinfectants". G. Wirtanen and S. Salo. 2003.]]] | ||
| − | Sodium hydroxide (caustic), [https://en.wikipedia.org/wiki/Ethylenediaminetetraacetic_acid EDTA (ethylene diaminetetra-acetic acid)], chlorinated disinfectants, and hydrogen peroxide-based disinfectants such as Pur-Ox from Birko or Lerasept-O from Loeffler are effective at breaking up biofilms when used in their highest recommended concentrations <ref name="Wirtanen_2001" /><ref>Brandon Jones. Private correspondence with Dan Pixley. 04/02/2018.</ref><ref>[https://www.reddit.com/r/TheBrewery/comments/6hqnvf/mtkettle_cleaning/dj0zd0s/ Levader on Reddit.com. "The Brewery". Retrieved 04/02/2018.]</ref>. Foaming agents that are often used in packaging lines for cleaning, however, might not be as effective. One study found that one foaming agent (VK10 Shureclean, which is sodium alkylbenzenesulphonate) required two times the maximum concentration that is recommended by the manufacturer to completely remove biofilms. In comparison, all of the sodium hydroxide (caustic) based cleaners that were tested were effective at completely removing biofilms in concentrations that were below the vendors' recommended maximum concentrations <ref>[https://link.springer.com/article/10.1007/s13213-010-0085-5#Bib1 Susceptibility of wine spoilage yeasts and bacteria in the planktonic state and in biofilms to disinfectants. Mariana Tristezza, António Lourenço, André Barata, Luísa Brito, Manuel Malfeito-Ferreira, Virgílio Loureiro. 2010.]</ref>. Peracetic acid (PAA) has also been shown to be effective against biofilms in the highest recommended concentrations but isn't as effective as the previously mentioned cleaners and should be used after a caustic cleaning cycle <ref>[https://www.researchgate.net/publication/273439407_Disinfectant_testing_against_brewery-related_biofilms Disinfectant testing against brewery-related biofilms. Storgårds, Erna & Närhi, Mikko & Wirtanen, Gun. 2001.]</ref><ref>[https://www.researchgate.net/publication/244994186_COMMERCIAL_SANITIZERS_EFFICACY_-_A_WINERY_TRIAL COMMERCIAL SANITIZERS EFFICACY – A WINERY TRIAL. Duarte, Filomena & López, Alberto & Alemão, Filomena & Santos, Rodrigo & Canas, Sara. 2011.]</ref>, but its effectiveness decreases below 20°C. Chlorine and iodine-based disinfectants destroy microbe at colder temperatures, however, they are less effective in the presence of wort or other residues. Chlorine-based disinfectants can cause pitting in stainless steel if left in contact for too long, and [https://ssbrewtech.zendesk.com/hc/en-us/articles/205602399-DO-NOT-USE-BLEACH-OR-CHLORINATED-CHEMICALS- some stainless steel manufacturers] recommend not using chlorine-based disinfectants at all (refer to your equipment and chemical manufacturers). Hot water is one of the most effective disinfectants, however, dry heat is not as effective at killing bacteria (one strain of ''L. brevis'' was able to withstand 80°C dry heat for 60 minutes) <ref name="Wirtanen_2001" />. Dry heat at higher temperatures will sterilize at 170°C for 1 hour or 190°C for 12 minutes and can be used to sterilize many metal and glass instruments. Flaming surfaces kills within seconds <ref>Private correspondence with Dr. Bryan Heit by Dan Pixley. 04/12/2018.</ref>. | + | Sodium hydroxide (caustic), [https://en.wikipedia.org/wiki/Ethylenediaminetetraacetic_acid EDTA (ethylene diaminetetra-acetic acid)], chlorinated disinfectants, and hydrogen peroxide-based disinfectants such as Pur-Ox from Birko or Lerasept-O from Loeffler are effective at breaking up biofilms when used in their highest recommended concentrations <ref name="Wirtanen_2001" /><ref>Brandon Jones. Private correspondence with Dan Pixley. 04/02/2018.</ref><ref>[https://www.reddit.com/r/TheBrewery/comments/6hqnvf/mtkettle_cleaning/dj0zd0s/ Levader on Reddit.com. "The Brewery". Retrieved 04/02/2018.]</ref>. Foaming agents that are often used in packaging lines for cleaning, however, might not be as effective. One study found that one foaming agent (VK10 Shureclean, which is sodium alkylbenzenesulphonate) required two times the maximum concentration that is recommended by the manufacturer to completely remove biofilms. In comparison, all of the sodium hydroxide (caustic) based cleaners that were tested were effective at completely removing biofilms in concentrations that were below the vendors' recommended maximum concentrations <ref>[https://link.springer.com/article/10.1007/s13213-010-0085-5#Bib1 Susceptibility of wine spoilage yeasts and bacteria in the planktonic state and in biofilms to disinfectants. Mariana Tristezza, António Lourenço, André Barata, Luísa Brito, Manuel Malfeito-Ferreira, Virgílio Loureiro. 2010.]</ref>. Another study tested six different types of cleaners were tested to see how well they removed the biofilms: keytones + surfactant detergent, quaternary ammonia + surfactant detergent, sodium hydroxide (caustic soda), sodium carbonate (soda ash), sodium hydroxide + surfactant (alkaline detergent), and chlorine (sanitizer, not a detergent). They found that only caustic soda was consistently efficient at removing the biofilm (the caustic, ammonia + surfactant cleaner, and the quaternary ammonia + surfactant detergent all efficiently removed cells that were adhered to a surface over 6 hours and were determined to be good for stopping biofilm formation from occurring in the first place) <ref>[https://www.researchgate.net/publication/235411588_Adhesion_and_biofilm_production_by_wine_isolates_of_Brettanomyces_bruxellensis Adhesion and biofilm production by wine isolates of Brettanomyces bruxellensis. C. M. Lucy Joseph, Gagandeep Renuka Kumar, Gagandeep Renuka Kumar, Edward Su, Linda F Bisson. 2006. American Journal of Enology and Viticulture 58(3):373-378.]</ref>. Peracetic acid (PAA) has also been shown to be effective against biofilms in the highest recommended concentrations but isn't as effective as the previously mentioned cleaners and should be used after a caustic cleaning cycle <ref>[https://www.researchgate.net/publication/273439407_Disinfectant_testing_against_brewery-related_biofilms Disinfectant testing against brewery-related biofilms. Storgårds, Erna & Närhi, Mikko & Wirtanen, Gun. 2001.]</ref><ref>[https://www.researchgate.net/publication/244994186_COMMERCIAL_SANITIZERS_EFFICACY_-_A_WINERY_TRIAL COMMERCIAL SANITIZERS EFFICACY – A WINERY TRIAL. Duarte, Filomena & López, Alberto & Alemão, Filomena & Santos, Rodrigo & Canas, Sara. 2011.]</ref>, but its effectiveness decreases below 20°C. Chlorine and iodine-based disinfectants destroy microbe at colder temperatures, however, they are less effective in the presence of wort or other residues. Chlorine-based disinfectants can cause pitting in stainless steel if left in contact for too long, and [https://ssbrewtech.zendesk.com/hc/en-us/articles/205602399-DO-NOT-USE-BLEACH-OR-CHLORINATED-CHEMICALS- some stainless steel manufacturers] recommend not using chlorine-based disinfectants at all (refer to your equipment and chemical manufacturers). Hot water is one of the most effective disinfectants, however, dry heat is not as effective at killing bacteria (one strain of ''L. brevis'' was able to withstand 80°C dry heat for 60 minutes) <ref name="Wirtanen_2001" />. Dry heat at higher temperatures will sterilize at 170°C for 1 hour or 190°C for 12 minutes and can be used to sterilize many metal and glass instruments. Flaming surfaces kills within seconds <ref>Private correspondence with Dr. Bryan Heit by Dan Pixley. 04/12/2018.</ref>. |
| + | |||
| + | Ethanol and isopropyl alcohol have also been found to be very effective sanitizers. The downside to these sanitizers is that they are flammable and are very volatile, evaporating quickly after use. One experiment by [http://masterbrewerspodcast.com/096-efficacy-of-sanitizers-in-the-brewery Elliot Parcells & Josh Pohlmann from Bells Brewery] claimed that iodophor was an ineffective sanitizer at concentrations of 25 ppm, however, their results have been questioned due to their methodology <ref>[https://www.facebook.com/groups/MilkTheFunk/permalink/2172245389470330/?comment_id=2172685769426292&comment_tracking=%7B%22tn%22%3A%22R%22%7D Dr. Bryan Heit. Milk The Funk Facebook thread on an MBAA podcast about the efficacy of iodophor as a sanitizer. 07/10/2018.]</ref>. | ||
| + | |||
| + | Sulfur dioxide has been used as a sanitizer in some cases in winemaking; see [https://www.facebook.com/groups/MilkTheFunk/permalink/2202288736465995/ this MTF thread] for more information. SO<sub>2</sub> is also used as a microbial stabilizer in finished wine. 50 ppm of total SO<sub>2</sub> can inhibit the growth of lactic acid bacteria in finished wine <ref>[https://psuwineandgrapes.wordpress.com/2018/10/26/understanding-difficult-malolactic-fermentations/ Dr. Molly Kelly. "Understanding Difficult Malolactic Fermentations". Wine & Grapes U. 10/26/2018. Retrieved 11/22/2018.]</ref>. 2.1 mg/L of molecular SO<sub>2</sub> is needed to inhibit ''Brettanomyces'' in finished wine <ref name="Agnolucci_2017">[https://link.springer.com/article/10.1007/s11274-017-2345-z Brettanomyces bruxellensis yeasts: impact on wine and winemaking. Monica Agnolucci, Antonio Tirelli, Luca Cocolin, Annita Toffanin. 2017.]</ref>. See [https://grapesandwine.cals.cornell.edu/newsletters/appellation-cornell/2012-newsletters/issue-12/article-contains-sulfites/ this article from Cornell] on an explanation between total SO<sub>2</sub>, free SO<sub>2</sub>, and bound SO<sub>2</sub>. | ||
| + | |||
| + | Caustic cleaners can errode glass when the pH gets above 9.0 <ref>[http://www.vitroglazings.com/VitroGlazings/media/sitedocuments/TDs/19_TD105F.pdf "How to Prevent Glass Corrosion". Paul F. Duffer. Glass Digest, November 15, 1986.]</ref><ref>[https://www.facebook.com/groups/MilkTheFunk/permalink/2223548741006661/?comment_id=2223604231001112&reply_comment_id=2224717294223139&comment_tracking=%7B%22tn%22%3A%22R%22%7D Shawn Savuto. Milk The Funk Facebook group post on caustic erroding glass. 08/12/2018.]</ref>. | ||
| + | |||
| + | Enzymes have been reported as a solution to helping to clean biofilms from within dispensing tubing. Dispensing line cleaners have been reported to be inneffective at cleaning established biofilms due to the protective exopolysaccharide (EPS) layer of biofilms and the proteins that yeast cells excrete during flocculation, which help them adhere to each other and to surfaces. Polysaccharases, polysaccharide lyases, and to a lesser extent, proteases, will catalyze EPS and proteins. Microbes secrete these enzymes themselves in order to free themselves from the biofilm when their environment becomes stressful, and the effect of a particular enzyme degrading a particular EPS depends on other EPS types in the biofilm; degrading one EPS type can lead to the growth of another species present in the biofilm. Thus, the effectiveness of enzyme cleaning depends on the enzymes used and the types of microbes in the biofilm. This is one potential problem with using enzymes for cleaning <ref name="Walker_2007">[https://onlinelibrary.wiley.com/doi/pdf/10.1002/j.2050-0416.2007.tb00257.x Removal of Microbial Biofilms from Dispense Equipment: The Effect of Enzymatic Pre-digestion and Detergent Treatment. Samantha L. Walker, Michail Fourgialakis, Belen Cerezo, and Stephen Livens. 2007.]</ref>. | ||
| + | |||
| + | For example, Walker et al. (2007) found that a mix of the enzymes alpha-amylase, beta-glucuronidase, glycose oxidase, dextrinase, protease, and pectinase followed by the use of a line cleaning agent reduced the number of viable ''S. cerevisiae'', ''Lactobacillus brevis'', and ''Leuconostoc mesenteroides'' by 2 logs, but the number of ''Flavimonas oryzihabitans'' increased. A doubling of the enzyme content resulted in more effectively elimating ''Flavimonas oryzihabitans'', but it still wasn't as effective as elimating the other microorganisms. Additionally, the enzymes were less effective on dispensing lines than they were on stainless steel. On dispensing lines, the bacteria were reduced, but yeast was not. The study concluded that a broad range of enzymes should be used, and might assist with breaking up biofilms when used in conjunction with line cleaning agents, but this depends on the types of microbes/EPS in the biofilm and some microbes might be more resistant to this treatment than others <ref name="Walker_2007" />. | ||
| − | + | ===Homebrew cleaners and disinfectants=== | |
'''Five Star Star San''' | '''Five Star Star San''' | ||
| − | Five Star Chemicals product Star San is a popular acid anionic sanitizer sold to homebrewers because of its relative safety and ease of use. Claims that acid anionic sanitizers are not effective at killing yeast have been made on various internet forums <ref>[https://www.homebrewersassociation.org/forum/index.php?topic=24447.msg312961#msg312961 User 'S. cerevisiae'. American Homebrewers Association forums. 10/05/2015. Retrieved 04/11/2018.]</ref><ref>[https://www.homebrewersassociation.org/forum/index.php?topic=4576.msg52894#msg52894 User 'richardt'. American Homebrewers Association forums. 11/15/2010. Retrieved 04/11/2018.]</ref>. These claims are based on the food science textbooks, "Principles of Food Sanitation," by Norman G. Marriott and Robert B. Gravani (2006) and "Basic Food Microbiology" by George Banward (1989), which contain conflicting information about the effectiveness of acid anionic sanitizers | + | Five Star Chemicals product Star San is a popular acid anionic sanitizer sold to homebrewers because of its relative safety and ease of use. Claims that acid anionic sanitizers are not effective at killing yeast have been made on various internet forums <ref>[https://www.homebrewersassociation.org/forum/index.php?topic=24447.msg312961#msg312961 User 'S. cerevisiae'. American Homebrewers Association forums. 10/05/2015. Retrieved 04/11/2018.]</ref><ref>[https://www.homebrewersassociation.org/forum/index.php?topic=4576.msg52894#msg52894 User 'richardt'. American Homebrewers Association forums. 11/15/2010. Retrieved 04/11/2018.]</ref><ref>[https://community.diybeer.com/topic/9709-starsan-not-effective-against-wild-yeast-and-molds/ Coopers Community Forums. Post by user 'Christinas1'. 09/08/2016. Retrieved 05/23/2019.]</ref>. These claims are cited in various food science pamphlets, blogs, and websites, and appear to be based on the food science textbooks, "Principles of Food Sanitation," by Norman G. Marriott and Robert B. Gravani (2006) and "Basic Food Microbiology" by George Banward (1989), which contain conflicting information about the effectiveness of acid anionic sanitizers. Neither of these textbooks contain experimental data nor references to experimental data. The statements in these books conflict with each other. Marriott and Gravani claim, "(Acid anionic sanitizers) have limited and varied antimicrobial activity (including poor yeast and mold activity)... The antimicrobial effect of acid anionics appears to be through reaction of the surfactant, with positively charged bacteria by ionic attraction to penetrate cell walls and disrupt cellular function." Banward's claims of acid anionic sanitizers are, "Advantages: Active against a wide spectrum of microorganisms including thermodurics, controis phage and most yeast strains. Disadvantages: Slow activity against sporeformers, not effective in destruction of most spores." Furthermore, the provided explanation, which is that acid anionic sanitizers supposedly don't work effectively against yeast and molds is because acid anionic sanitizers are negatively charged and yeast are also negatively charged yet bacteria is killed because it is positively charged, is biologically incorrect ([https://etd.ohiolink.edu/pg_10?0::NO:10:P10_ACCESSION_NUM:osu1250193404 this masters thesis] appears to be the source of this incorrect information). According to [https://www.youtube.com/watch?v=0JC9n50RdVo Dr. Bryan Heit of Sui Generis blog], both yeast and bacteria have negatively charged cell walls, and this fact has been well established in microbiology since the 1940's (Dr. Heit has published several [https://scholar.google.com/citations?user=8yxqYNgAAAAJ&hl=en&oi=ao peer-reviewed scientific studies on cell wall polarity]). |
| − | While we are not aware of any publicly available published studies on the efficacy of StarSan, several studies with other acid anionic sanitizers have confirmed that they are effective against | + | Star San has only been officially tested by Five Star Chemicals against the pathogenic bacteria species ''E. coli'' and ''S. Aureus'', which is the minimum baseline required by the EPA to be labeled a "sanitizer" <ref>[https://www.homebrewersassociation.org/forum/index.php?topic=31537.msg409346#msg409346 Conn, Denny. American Homebrew Association forums. 04/08/2018. retrieved 07/09/2018.]</ref>. While we are not aware of any publicly available published studies on the efficacy of StarSan to kill yeast, several studies with other acid anionic sanitizers have confirmed that they are effective against yeast. Lee et al. (2007) found that an acid sanitizer very similar to Star San that uses citric acid instead of phosphate but the same surfactant (sodium dodecylbenzene sulfonate) took 5 minutes to kill ''Saccharomyces cerevisiae'', ''E. coli'', and ''Listeria innocua'' at room temperature (some species were killed faster than others with the ''E. coli'' actually being more resistant than the yeast), and one minute if the sanitizer was heated to 40°C on both metal and LDPE plastic (they compared the acid anionic sanitizer to 35% hydrogen peroxide, which killed all organisms with 15 seconds, indicating that this acid anionic sanitizer is effective at killing yeast, but it takes longer than a stronger chemical such as hydrogen peroxide). This study did not make mention of biofilms, however, the cultures were allowed to grow and dry overnight which could have allowed for biofilm formation <ref>[https://onlinelibrary.wiley.com/doi/full/10.1111/j.1750-3841.2007.00496.x Efficacy of Two Acidic Sanitizers for Microbial Reduction on Metal Cans and Low-Density Polyethylene Film Surfaces. J. LEE, M.J. GUPTA, J. LOPES, AND M.A. PASCALL. 2007.]</ref>. Five Star Chemicals [https://bsgcraft.com/Resources/Craftbrewing/PDFs/Product_Spec_and_Data_Sheets/Product_Spec_Sheets/StarSan_Tech.pdf also recommends 5 minutes of contact time with Star San]. Winniczuk et al. (1997) found that three phosphoric acid anionic sanitizers ("CS-100" and "CS-101-lf" by Chemical Systems of Florida, and "Clear-Clean" by Pelican Brand) were less effective at killing yeast than bacteria in the timeframe tested (1 minute contact time), but they were still effective at killing yeast at high concentrations (peracetic acid also required a higher concentration to kill yeast than bacteria). However, one of the acid anionic sanitizers tested was more effective than the other two, indicating that the chemical makeup of the particular acid anionic sanitizer has an impact on how effective it is as a sanitizer relative to other acid anionic sanitizers. Additionally, they found that peracetic acid, iodophor, and chlorine dioxide required less concentration than the acid anionic sanitizers to be effective (again, tested at 1 minute exposure time) <ref>[http://lp7lc5er8n.scholar.serialssolutions.com/?sid=google&auinit=PP&aulast=Winniczuk&atitle=Minimum+inhibitory+concentrations+of+antimicrobials+against+micro-organisms+related+to+citrus+juice&id=doi:10.1006/fmic.1997.0103&title=Food+microbiology&volume=14&issue=4&date=1997&spage=373&issn=0740-0020 Minimum inhibitory concentrations of antimicrobials against micro-organisms related to citrus juice. P.P Winniczuk, M.E Parish. 1997.]</ref>. |
| − | Reduction on Metal Cans and Low-Density Polyethylene Film Surfaces. J. | ||
See [https://www.facebook.com/groups/MilkTheFunk/permalink/1436419659719577/ this MTF thread] for a more extensive explanation of why skepticism should be applied to the claim that acid anionic sanitizers are not effective at killing yeast. | See [https://www.facebook.com/groups/MilkTheFunk/permalink/1436419659719577/ this MTF thread] for a more extensive explanation of why skepticism should be applied to the claim that acid anionic sanitizers are not effective at killing yeast. | ||
| Line 90: | Line 170: | ||
# Leave in contact with surfaces for 5 minutes or more. | # Leave in contact with surfaces for 5 minutes or more. | ||
# Optional: warm the Star San and water solution to 40°C/104°F. | # Optional: warm the Star San and water solution to 40°C/104°F. | ||
| + | |||
| + | '''Iodophor''' | ||
| + | |||
| + | A popular alternative to Star San is iodophor, which is an iodine-based sanitizer. | ||
| + | |||
| + | [http://masterbrewerspodcast.com/096-efficacy-of-sanitizers-in-the-brewery Elliot Parcells & Josh Pohlmann from Bells Brewery] tested the efficiency of various sanitizers to kill ''Lactobacillus'', ''Brettanomyces'', and diastatic strains of ''Saccharomyces cerevisiae''', and claimed that iodophor was ineffective at concentrations of 25 ppm, which is the maximum concentration of iodophor for it to be considered a no-rinse sanitizer. The methodology of this experiment has [https://suigenerisbrewing.com/index.php/2022/01/20/how-not-to-test-a-sanitizer/ received some criticism] that might invalidate its results. They diluted yeast/bacteria cultures on a 1:10 ratio into iodophor, thus diluting the iodophor below its recommended dilution and exposing it to concentrations of microbes far beyond what it was designed to kill. Testing the ability to sterilize a bulk solution does not translate to the ability of the sanitizer to sanitize trace surface microbes after proper cleaning <ref>[https://www.facebook.com/groups/MilkTheFunk/permalink/2172245389470330/?comment_id=2172685769426292&comment_tracking=%7B%22tn%22%3A%22R%22%7D Dr. Bryan Heit. Milk The Funk Facebook thread on an MBAA podcast about the efficacy of iodohor as a sanitizer. 07/10/2018.]</ref>. | ||
'''Five Star PBW''' | '''Five Star PBW''' | ||
| − | PBW is a replacement for caustic and is much safer to use and dispose of. It is non-corrosive and safe to use on stainless steel and plastics. We are not aware of any studies that examine the efficacy of P.B.W. to remove biofilms. | + | PBW is a replacement for caustic and is much safer to use and dispose of. It is non-corrosive and safe to use on stainless steel and plastics. We are not aware of any studies that examine the efficacy of P.B.W. to remove biofilms, nor any other percarbonate-based <ref>[https://patents.google.com/patent/US5663132 "Non-caustic composition comprising peroxygen compound and metasilicate and cleaning methods for using same". US patent for PBW. 03/01/1995. Retrieved 04/19/2018.]</ref> cleaners efficacy in a brewing environment. |
Tips for using PBW on equipment that may have had biofilm formation <ref>[http://www.fivestarchemicals.com/wp-content/uploads/PBWTech.pdf "PBW Tech Sheet". Five Star Chemicals. Retrieved 04/19/2018.]</ref>: | Tips for using PBW on equipment that may have had biofilm formation <ref>[http://www.fivestarchemicals.com/wp-content/uploads/PBWTech.pdf "PBW Tech Sheet". Five Star Chemicals. Retrieved 04/19/2018.]</ref>: | ||
| − | # For circulated cleaning, use 3 ounces per gallon | + | # For circulated cleaning, use 1 to 3 ounces per gallon depending on the soil load, and heat to 180°F. Recirculate for 30 minutes (if using this product on plastics, heat the water that is mixed with the PBW to be as hot as the manufacturer of the plastics recommends, and extend the circulation time to an hour or two). |
| − | # For non-circulated | + | # For non-circulated, soaking, use 6 to 8 ounces per gallon of 180°F water (if using this product on plastics, heat the water that is mixed with the PBW to be as hot as the manufacturer of the plastics recommends), and soak overnight. Note that there have been reports of high concentrations of PBW breaking down PET bottles over time when soaking <ref>[https://www.homebrewtalk.com/forum/threads/pbw-and-plastics.521708/ Homebrewtalk thread reporting PBW breaking down PET bottles. 03/25/2013.]</ref>. For example, BetterBottle™ recommends using 5 grams per liter of 125°F water (0.67 ounces per gallon, or one tablespoon per gallon) of PBW and cleaned with agitation/circulation instead of soaking. They also recommend using an enzymatic cleaner such as Seventh Generation Free and Clear Natural 2X or Super Pro-zyme Enzymatic Cleaner instead of PBW <ref>[https://www.facebook.com/groups/MilkTheFunk/permalink/2104526149575588/ Better Bottle Cleaning and Sanitizing web article; downloaded from Archive.org.]</ref>. |
# Rinse thoroughly with hot water (cold water may not remove some of the residual chemicals). | # Rinse thoroughly with hot water (cold water may not remove some of the residual chemicals). | ||
| + | |||
| + | Alternatives to PBW include: | ||
| + | * [https://www.nationalchemicals.com/craftmeister/ CraftMeister Alkaline Brewery Wash.] | ||
| + | * [https://www.google.com/search?client=firefox-b-1-d&q=pbw+vs+oxiclean Unscented Oxiclean.] | ||
| + | |||
| + | '''See Also''' | ||
| + | * [http://realbeer.com/jjpalmer/cleaning.html "A Complete Guide to Cleaning and Sanitation" by John Palmer.] | ||
| + | * "How To Brew". Chapter 2 Cleaning & Sanitizing, by John Palmer, 4th Edition, 2017. | ||
| + | * [https://www.facebook.com/groups/MilkTheFunk/permalink/2104526149575588/ BetterBottle Guide to cleaning and sanitizing PET.] | ||
==Quality Control== | ==Quality Control== | ||
(To do) | (To do) | ||
| − | '''Quality Control''' is the process of identifying quality problems in the product, and is a reactive process aimed at correcting a detected problem <ref name="Diffen" />. | + | ===Quality Control=== |
| + | '''Quality Control''' is the process of identifying quality problems in the product, and is a reactive process aimed at correcting a detected problem <ref name="Diffen" />. | ||
| − | Detection methods <ref name="Wirtanen_2001" />: | + | Detection methods <ref name="Wirtanen_2001" /><ref name="Bokulich_2018" />: |
| − | * [http://cdn.intechopen.com/pdfs/27440/InTech-Use_of_atp_bioluminescence_for_rapid_detection_and_enumeration_of_contaminants_the_milliflex_rapid_microbiology_detection_and_enumeration_system.pdf ATP bioluminescence system monitors for contaminants.] | + | * [http://cdn.intechopen.com/pdfs/27440/InTech-Use_of_atp_bioluminescence_for_rapid_detection_and_enumeration_of_contaminants_the_milliflex_rapid_microbiology_detection_and_enumeration_system.pdf ATP bioluminescence system monitors for contaminants.] This method uses light to detect organic matter, but cannot identify the microorganisms that are present. It also indicates whether the biological matter is low enough to employ disinfectant. |
* Swabbing and cultivating on agar and PCR. | * Swabbing and cultivating on agar and PCR. | ||
* Direct epifluorescence filter technique (DEFT)/microcolony method. | * Direct epifluorescence filter technique (DEFT)/microcolony method. | ||
* Direct impedimetry. | * Direct impedimetry. | ||
* Flow cytometry. | * Flow cytometry. | ||
| + | * DGGE/TGGE (separation of heterologous sequences by chemical/heat denaturation). | ||
| + | * Clone Libraries (Sanger sequencing of transformed ''E. coli'' clones containing diagnostic gene amplicons). | ||
| + | * qPCR | ||
| + | * [http://illumina.com/ Illumina] Sequencing | ||
| + | * 454 Life Sciences Pyrosequencing | ||
| + | * [https://europepmc.org/abstract/med/29703269 PCR DNA dipstick for rapid detection at different stages of the brewing process]. | ||
| + | * Disposable electrochemical biosensors for ''Brettanomyces bruxellensis'' in wine <ref>[https://www.sciencedirect.com/science/article/pii/S0925400518317271 Disposable electrochemical biosensors for Brettanomyces bruxellensis and total yeast content in wine based on core-shell magnetic nanoparticles. María L. Villalong, Boryana Borisova, Christian B. Arenas, Anabel Villalonga, María Arévalo-Villena, Alfredo Sánchez, José M. Pingarrón, Ana Briones-Pérez, Reynaldo Villalong. 2018.]</ref> | ||
| + | * [https://www.scribd.com/document/395419129/Micro-Identification?secret_password=yFUCiiLek4p72XWmAzMK&fbclid=IwAR1y4-yrXgpoqtyPsSTt8scfM41OBMX3hhRi0InSlLUNIIeRA8yS-3ezs_k PDF from Richard Preiss with a summary mostly derived from Jorgenson, E. (2017). An Overview of Bacteria Found in Brewing Ecosystems. Master Brew. Assoc.] | ||
| + | * [https://onlinelibrary.wiley.com/doi/pdf/10.1111/1751-7915.13422 RNA-FISH (Fluorescense ''In Situ'' Hybridization) technique for detecting ''Brettanomyces'' contamination should use a red-emitting fluorophore.] | ||
| + | * [https://www.sciencedirect.com/science/article/pii/S0030402619318546 Surface enhanced Raman spectroscopy phylogenetictree for genosensing of ''Brettanomyces bruxellensis'' yeast on nanostructured ultrafine glass supports.] | ||
| − | Beer adapted strains of ''Brettanomyces'' and lactic acid bacteria have been found to be more difficult to culture on agar than when they are not adapted to the environment. Malt agar has been shown to be more effective at showing growth than other types of agar (Dekkera Medium, Universal Beer Agar, Potato Dextrose Agar, and Beer Agar). [[Laboratory_Techniques#Saccharomyces|YPD]] with 10 ppm Cycloheximide was not tested in the study, but Nick Impellitteri, the owner of [[The Yeast Bay]], reports that this media works well for him <ref>[https://www.facebook.com/groups/MilkTheFunk/permalink/2047731471921723/?comment_id=2047735648587972&reply_comment_id=2047763545251849&comment_tracking=%7B%22tn%22%3A%22R0%22%7D Nick Impellitteri. Milk The Funk Facebook post on the ability of different media to grow beer adapted ''Brettanomyces''. 04/03/2018.]</ref>, and DBDM media has also been reported to work well for growing ''Brettanomyces''. In addition, it is recommended that 100 mL samples are taken since beer adapted contaminants are harder to grow on media, and at least 7 days of incubation time should be allowed for ''Brettanomyces'' to show signs of growth <ref>[https://www.tandfonline.com/doi/abs/10.1094/ASBCJ-2008-0917-01 Effects of Beer Adaptation on Culturability of Beer-Spoilage Dekkera/Brettanomyces Yeasts. Koji Suzuki, Shizuka Asano, Kazumaru Iijima, Tomoo Ogata, Yasushi Kitagawa & Tsunehiro Ikeda. 2018.]</ref> (see also [https://www.facebook.com/groups/MilkTheFunk/permalink/2047731471921723/ this MTF thread)]. | + | Beer adapted strains of ''Brettanomyces'' and lactic acid bacteria have been found to be more difficult to culture on agar than when they are not adapted to the environment <ref name="Suzuki_2012" />. Malt agar has been shown to be more effective at showing growth than some other types of agar (Dekkera Medium, Universal Beer Agar, Potato Dextrose Agar, and Beer Agar). [[Laboratory_Techniques#Saccharomyces|YPD]] with 10 ppm Cycloheximide was not tested in the study, but Nick Impellitteri, the owner of [[The Yeast Bay]], reports that this media works well for him <ref>[https://www.facebook.com/groups/MilkTheFunk/permalink/2047731471921723/?comment_id=2047735648587972&reply_comment_id=2047763545251849&comment_tracking=%7B%22tn%22%3A%22R0%22%7D Nick Impellitteri. Milk The Funk Facebook post on the ability of different media to grow beer adapted ''Brettanomyces''. 04/03/2018.]</ref>, and DBDM media has also been reported to work well for growing ''Brettanomyces''. In addition, it is recommended that 100 mL samples are taken since beer adapted contaminants are harder to grow on media, and at least 7 days of incubation time should be allowed for ''Brettanomyces'' to show signs of growth <ref>[https://www.tandfonline.com/doi/abs/10.1094/ASBCJ-2008-0917-01 Effects of Beer Adaptation on Culturability of Beer-Spoilage Dekkera/Brettanomyces Yeasts. Koji Suzuki, Shizuka Asano, Kazumaru Iijima, Tomoo Ogata, Yasushi Kitagawa & Tsunehiro Ikeda. 2018.]</ref> (see also [https://www.facebook.com/groups/MilkTheFunk/permalink/2047731471921723/ this MTF thread)]. |
| − | Advanced beer-spoiler detection medium (ABD) has been shown as a more effective growth medium compared to other media when attempting to grow beer-adapted bacteria such as hop tolerant strains of ''Lactobacillus'' that don't grow well on other media. | + | Advanced beer-spoiler detection medium (ABD) has been shown as a more effective growth medium compared to other media when attempting to grow beer-adapted bacteria such as hop tolerant strains of ''Lactobacillus'' that don't grow well on other media <ref name="Suzuki_2012" />. |
| + | |||
| + | See also: | ||
| + | * [[Laboratory Techniques]] for the recipes for these agar types. | ||
| + | *[https://www.brewersassociation.org/directories/suppliers Brewers Association list of lab services (filter by "Services", then "Contamination" or "Laboratory services").] | ||
| + | * [https://drive.google.com/drive/folders/1IsB_deiFyJfwKbHiKFf5-YCWwEufHQk1 Kevin McGabe's HomebrewCon 2018 Seminar "Bringing a Brewery Quality Control Lab Into Your Home".] | ||
| + | * [https://www.mbaa.com/publications/tq/tqPastIssues/2017/Documents/TQ-54-2-0408-01.pdf MBAA guide to common spoilage bacteria and their morphology.] | ||
| + | * [https://www.mbaa.com/publications/tq/tqPastIssues/2020/Pages/TQ-57-1-0222-01.aspx MBAA TQ article, "Building a Quality Control Lab: An Introduction to Starting and Growing a Quality Program" by Melissa Antone.] | ||
| + | * [https://file.scirp.org/pdf/AiM_2016032215023444.pdf Review of Gram negative bacteria in brewing.] | ||
| + | * [http://suigenerisbrewing.com/index.php/2019/12/04/contamination-detection-3/ Dr. Bryan Heit of Sui Generis blog explains his PCR method for detecting diastatic yeast.] | ||
| + | * [https://www.facebook.com/groups/MilkTheFunk/posts/5310978085597029 Zach Taggart offers QC tips on testing ''Lactobacillus'' growth using modified MRS agar plating and recording morphology.] | ||
| + | * [https://www.masterbrewerspodcast.com/196 MBAA Podcast with Goose Island on how they traced an elusive wild yeast infection in their brewery.] | ||
| + | * Books: | ||
| + | ** [https://my.mbaa.com/ItemDetail?iProductCode=52206 "Illustrated Guide to Microbes and Sediments in Wine, Beer and Juice" by George G. Edwards.] | ||
| + | ** [https://www.carllibri.com/colouratlas "Colour Atlas and handbook of Beverage Biology" by Dr. Werner.] | ||
| + | |||
| + | ====Viable But Nonculturable==== | ||
| + | "Viable but nonculturable" ('''VBNC''') is a newly identified state for bacteria that are not able to grow or form colonies on typical growth media (i.e., lack of cell division), but they remain viable (alive) and retain a limited level of metabolic activity (reduced nutrient transport, respiration, and synthesis of compounds) while sometimes being able to regain their population when returned to a more ideal environment. The cells often exhibit dwarfing, and they can remain in this state without dying for 4-12+ months depending on the species. An appropriate viability test for a given species can be performed to show that the cells are not dead, even though they don't grow on typical growth media (for example, intracellular hydrolysis of CTC or reduction of INT as an indication of metabolic activity, by establishing the presence of an intact cytoplasmic membrane via BacLight® or propidium iodide, or by multi-parameter flow cytometry). Cells enter this state as a way to survive some sort of stress in their environment (for example, osmotic stress, too much oxygen exposure, exposure to white light, etc.). Treatments such as pasteurization in milk and chlorination of wastewater have also been shown to induce VBNC. A number of species have been found to be able to enter the VBNC state, including ''E. coli'', ''Lactobacillus plantarum'', ''L. lactis'', ''L. linderi'', ''L. casei'', ''L. plantarum'', ''L. paracollinocides'', ''L. acetotolerans'', and several species of ''Salmonella''. Early studies on VBNC microbes were not able to fully show that the resuscitation was truly from VBNC cells rather than a very small number of culturable cells, but later studies were able to show that some bacteria can be resuscitated from a VBNC state, although most bacteria that enter a VBNC state have not been shown to be able to be resuscitated <ref>[https://pdfs.semanticscholar.org/e661/934dca6bb0dbb31a8781f3193232b7b5a8a4.pdf The Viable but Nonculturable State in Bacteria. James D. Oliver. The Journal of Microbiology. 2005.]</ref><ref name="Liu_2018">[https://www.frontiersin.org/articles/10.3389/fmicb.2018.02076/full#B28 Induction and Recovery of the Viable but Nonculturable State of Hop-Resistance Lactobacillus brevis. Junyan Liu, Yang Deng, Thanapop Soteyome, Yanyan Li, Jianyu Su, Lin Li, Bing Li, Mark E. Shirtliff, Zhenbo Xu, and Brian M. Peters. Front. Microbiol. 2018. DOI: https://doi.org/10.3389/fmicb.2018.02076.]</ref>. The concept of VBNC cells is somewhat controversial in microbiology; some experts argue that there is no difference between so-called "VBNC" cells and persister cells <ref>[https://sfamjournals.onlinelibrary.wiley.com/doi/abs/10.1111/1462-2920.15463 Song, S. and Wood, T.K. (2021), ‘Viable but non-culturable cells’ are dead. Environ Microbiol, 23: 2335-2338. https://doi.org/10.1111/1462-2920.15463.]</ref>. See also this [https://www.facebook.com/groups/MilkTheFunk/posts/6036200423074788/ MTF thread by Dr. Bryan Heit]. | ||
| + | |||
| + | A couple of published studies have reported inducing the VBNC state in bacteria strains that were isolated from contaminated beer. Liu et al. (2017) were able to induce a VBNC state (meaning that they were not able to grow on growth media for up to 14 days) in a strain of ''Lactobacillus lindneri'', a species that is responsible for 15-25% of spoiled beer reports, and determined VBNC via a Live/Dead BacLight® bacterial viability kit. They induced this state by storing the cells in beer at 0°C without shaking for 190 days. They also found that storing the VBNC cells at -80°C in glycerol stocks was the best way to maintain the cells. They were able to resuscitate the cells by growing them on MRS media that had 500-1000-μL of the enzyme catalase spread onto them (trying higher temperatures did not work to resuscitate, nor using higher concentrations of MRS), thus showing that brewers can use catalase to help grow VBNC state ''L. lindneri'' cells on MRS media (and perhaps other species of ''Lactobacillus'' as well). It took 3-4 days to begin showing signs of growth on the catalase supplemented MRS media. It was also demonstrated that VBNC cells could grow in beer after 30 days of incubation, and showed final cell counts similar to normal ''L. lindneri'' and resuscitated cells <ref>[https://www.sciencedirect.com/science/article/pii/S0882401017303030?via%3Dihub First study on the formation and resuscitation of viable but nonculturable state and beer spoilage capability of Lactobacillus lindneri. Junyan Liu, Lin Lia, Bing Li, Brian M. Peters, Yang Deng, Zhenbo Xua, Mark E. Shirtliff. Microbial Pathogenesis, Vol 107. 2017. DOI: https://doi.org/10.1016/j.micpath.2017.03.043.]</ref>. Lui et al. (2018) reproduced these results with a hop tolerant strain of ''L. brevis'' <ref name="Liu_2018" />. The genes that are associated with VBNC were also found in a beer contaminating strain of ''Lactobacillus acetolerans'' <ref>[https://www.tandfonline.com/doi/abs/10.1080/03610470.2021.1997280 Chunguang Luan, Weihua Cao, Na Luo, Jingxia Tu, Jianqin Hao, Yihong Bao, Feike Hao, Deliang Wang & Xin Jiang (2021) Genomic Insights into the Adaptability of the Spoilage Bacterium Lactobacillus acetotolerans CN247 to the Beer Microenvironment, Journal of the American Society of Brewing Chemists, DOI: 10.1080/03610470.2021.1997280.]</ref>. | ||
| + | |||
| + | See also: | ||
| + | * [[Pediococcus#VBNC|VBNC in ''Pediococcus'']] | ||
| + | |||
| + | =====VBNC In Yeast===== | ||
| + | While the VBNC state has mostly only been studied in detail for bacteria, it has also been suggested that this state is also possible with eukaryotes (yeast). It has been reported that although there are many methods for detecting ''Brettanomyces'' in winemaking, there are cases when ''Brettanomyces'' is not found using culturing techniques, but years later have still infected wine. Agnolucci et al. (2010) found that sulfur dioxide induces the VBNC state in 7 ''Brettanomyces'' strains isolated from wine at concentrations of 0.2 mg/L (molecular SO<sub>2</sub>) for 5 out of the 7 strains to 0.4 mg/L for the other 2 strains after 24 hours of incubation in a synthetic wine medium that was supplemented with various amounts of sulfur dioxide. At 0.4 mg/L they found that all but one strain had 0% culturable cells, and 4-26% VBNC cells depending on the strain. At 0.8 mg/L, no strains had culturable cells, but they all had at least 4.6-17% VBNC cells (percent of the original number of cells before being exposed to the sulfur dioxide). Even at 1 mg/L of sulfur dioxide levels, there were 2.9-15% VBNC cells, depending on the strain (the ability for the ''Brettanomyces'' to remain viable at 1mg/L of sulfur dioxide might have also been due to the pH only being 3.5, and ethanol only being 13%). They also found that 2.1 mg/L was required to reduce the VBNC state of cells to zero after 55 days of incubation and limit the amount of ethyl phenols produced by ''Brettanomyces''. They reported that trypan blue was the best method for detecting VBNC cells <ref>[https://www.sciencedirect.com/science/article/pii/S0168160510003958?via%3Dihub Sulphur dioxide affects culturability and volatile phenol production by Brettanomyces/Dekkera bruxellensis. Agnolucci M, Rea F, Sbrana C, Cristani C, Fracassetti D, Tirelli A, Nuti M. 2010. DOI: https://doi.org/10.1016/j.ijfoodmicro.2010.07.022.]</ref>. | ||
| + | |||
| + | The Agnolucci et al. (2010) study did not provide a method or data for resuscitating the VBNC ''Brettanomyces'' cells so that they can again divide and grow colonies, and this has been criticized because resuscitation of VBNC cells is considered an important aspect of strengthening the conclusion that the cells are indeed VBNC. Serpaggi et al. (2012) found similar results with using 0.8 mg/L of molecular SO<sub>2</sub>, which resulted in all ''Brettanomyces'' cells from one wine strain to not be culturable on YPD after being incubated in synthetic wine medium that was supplemented with SO<sub>2</sub> for 2 days, while the viability of the cells remained high for as long as 11 days (they did not check viability after 11 days, and the viability count remained constant from day 2 to day 11; viability was determined by staining with fluorescein diacetate which stains when certain metabolic esterase activity is present). They were able to resuscitate the cells by adding NaOH to the media to bring the pH up from 3.5 to 4.0 in order to effectively eliminate the molecular sulfur dioxide (molecular SO<sub>2</sub> is the only form of SO<sub>2</sub> that is significantly effective at inhibiting microbes, and it is only stable at very low pH's). They noted that VBNC cells were about 20% smaller in size than culturable cells. The cells in the VBNC state did not produce 4EG phenol but did produce a very small amount of 4EP phenol <ref>[https://www.ncbi.nlm.nih.gov/pubmed/22365358 Characterization of the "viable but nonculturable" (VBNC) state in the wine spoilage yeast Brettanomyces. Serpaggi V, Remize F, Recorbet G, Gaudot-Dumas E, Sequeira-Le Grand A, Alexandre H. 2012. DOI: 10.1016/j.fm.2011.12.020.]</ref>. It has also been demonstrated that the presence of minerals and vitamins, as well as p-coumaric acid, can assist in resuscitating so-called VBNC cells of ''Brettanomyces'' <ref>[https://www.mdpi.com/2306-5710/9/3/69 Chandra M, Branco P, Prista C, Malfeito-Ferreira M. Role of p-Coumaric Acid and Micronutrients in Sulfur Dioxide Tolerance in Brettanomyces bruxellensis. Beverages. 2023; 9(3):69. https://doi.org/10.3390/beverages9030069.]</ref>. | ||
| + | |||
| + | It is worth noting that the VBNC state in ''Brettanomyces'' has not been tested against a higher concentration of SO<sub>2</sub> (for example when wineries use much higher concentrations of SO<sub>2</sub> solutions as a sanitizer), other chemical sanitizers, and pasteurization-level temperatures. For example, it has been proposed that steaming barrels in order for them to reach 140°F (60°C) for 20 minutes is enough to sanitize them (see [[Barrel#Sanitizing|Barrel Sanitizing]] for more information). However, Nunes de Lima et al. (2020) inoculated wines with various strains of ''Brettanomyces'' after raising the pH of the wine to 3.8, which rendered the amount of free SO<sub>2</sub> in the wines completely ineffective, and many of the strains entered a VBNC state. This indicates that other unknown factors can induce a VBNC state in ''Brettanomyces''. It has been documented that other factors, such as nutrient starvation, extreme temperatures, osmotic pressure and oxygen, has caused a VBNC state in other microorganisms (VBNC has been mostly studied in bacteria) <ref name="Nunes de Lima 2020">[https://www.sciencedirect.com/science/article/abs/pii/S0740002020302069 Survival and metabolism of hydroxycinnamic acids by Dekkera bruxellensis in monovarietal wines. Adriana Nunes de Lima, Rui Magalhães, Francisco Manuel Campos, José António Couto. 2020. DOI: https://doi.org/10.1016/j.fm.2020.103617.]</ref>. | ||
| + | |||
| + | ===Sensory Analysis=== | ||
| + | Sensory analysis is the process of implementing a standardized method for collecting data on finished beer through sensory evaluation. Sensory analysis can be a low tech approach to first steps in identifying potential quality issues <ref>[https://beerandbrewing.com/dictionary/nB20s1RI6j/ "Sensory Evaluation" Craft Beer & Brewing website. Retrieved 02/14/2020.]</ref>. | ||
| + | |||
| + | See also: | ||
| + | * [https://beerandbrewing.com/dictionary/nB20s1RI6j/ "Sensory Evaluation" on Craft beer & Brewing website.] | ||
| + | * [https://www.morebeer.com/articles/Beer_Sensory_Analysis "An Introduction to Sensory Analysis" by Scott Bickham on the Morebeer website.] | ||
| + | * [https://www.youtube.com/channel/UCql1o9GpzeDqWHgd52Zxm9Q BrewEssence product videos.] | ||
| − | + | ===Supply Chain Management=== | |
| + | * [https://www.brewersassociation.org/presentations/supply-chain-management-for-craft-brewers/ Supply Chain Management on the BA Website.] | ||
==Quality Improvement== | ==Quality Improvement== | ||
| Line 126: | Line 270: | ||
==See Also== | ==See Also== | ||
===Additional Articles on MTF Wiki=== | ===Additional Articles on MTF Wiki=== | ||
| + | * [[Laboratory Techniques]] | ||
| + | * [[Wild_Yeast_Isolation#Safety|Wild Microbe Safety]] | ||
| + | * [[Mold]] | ||
| + | * [[Grain#Microbial_Populations_on_Barley|Microbial Populations on Grain and Malted Barley]] | ||
* [[Mixed Fermentation]] | * [[Mixed Fermentation]] | ||
| − | ===External Resources=== | + | ===External Resources=== |
| + | * [https://www.youtube.com/watch?v=TyBiDvphGHs Cleaning and Sanitation with Rick Theiner - BeerSmith Podcast #187.] | ||
| + | * [http://masterbrewerspodcast.com/115-todays-challenges-in-beer-quality MBAA Podcast interview with Mary Pellettieri, author of "Quality Management: Essential Planning for Breweries".] | ||
| + | * [https://www.masterbrewerspodcast.com/045 MBAA Podcast interview with Eric Jorgenson about his approach to microbiology and his quick reference guide of significant bacteria found in the brewery environment.] | ||
| + | * [https://www.masterbrewerspodcast.com/271 MBAA Podcast interview with Lauryn Rivera and Tess Downer about Comprehensive Quality at Odell's.] | ||
| + | * [https://www.masterbrewerspodcast.com/episodes MBAA Podcast interview with Jack Van Paepeghem about troubleshooting and identifying ''Pectinatus'' (viscous and hydrogen sulfide in lagers) contamination in a CFT 10 head 2 seamer.] | ||
| + | * "Quality Management: Essential Planning for Breweries" by Mary Pellettieri (Brewers Publications), 2015. | ||
| + | * "Illustrated Guide to Microbes and Sediments in Wine, Beer & Juice" by Charles G. Edwards (WineBuggs LLC), 2005 - A microscope companion book that includes over 30 different species of yeast, bacteria and mold commonly associated with beverages, as well as frequently encountered sediments. | ||
| + | * [http://my.asbcnet.org/ItemDetail?iProductCode=96346 "QUALITY SYSTEMS: Practical Guides for Beer Quality", by Dr. Charlie Bamforth, 2019.] | ||
| + | * [http://my.mbaa.com/ItemDetail?iProductCode=72697 "Brewery Cleaning: Equipment, Procedures, and Troubleshooting" by Richard J. Rench.] | ||
| + | * [https://jp.hach.com/asset-get.download.jsa?id=50544340479 Industry standards of dissolved oxygen levels in beer throughout the brewing process, by Hach.] | ||
| + | * [https://eurekabrewing.wordpress.com/yeast-cultivation/ Eureka Brewing Blog on lab techniques, yeast handling, etc.] | ||
| + | * "Yeast: A Practical Guide to Beer Fermentation" by Chris White and Jamil Zainesheff, 2010. | ||
| + | * "Brewing Yeast and Fermentation" by Boulton and Quain, 2006. | ||
| + | * [https://www.asbcnet.org/Pages/default.aspx American Society of Brewing Chemists.] | ||
| + | * [https://brewingscience.com/brewers-lab-handbook/ "Brewers' Laboratory Handbook: Brewing Without the Blindfold™", by Brewing Science Institute.] | ||
| + | * [https://bkyeast.wordpress.com/2012/02/19/selective-media-part-i/ "Selective Media Part 1" by BKYeast blog.] | ||
| + | * [https://bkyeast.wordpress.com/2013/05/06/pouring-plates-and-making-slants/ "Pouring Plates and Making Slants" by BKYeast blog.] | ||
| + | * [https://www.escarpmentlabs.com/single-post/2019/06/17/Quality-management-at-Escarpment-Labs Quality Management at Escarpment Labs.] | ||
| + | * [http://www.terifahrendorf.com/Lab-Manual.pdf Teri Fahrendorf’s "Small Brewery Lab Procedures Manual"] | ||
| + | * [https://coloradobeer.org/tech-safety-post/so-you-want-to-add-a-brewing-lab/ Colorado Brewers Guild "So You Want to Add a Brewing Lab?"] | ||
| + | * [https://www.youtube.com/watch?v=bW-UkOHtx9k Brew Strong podcast interview with Dr. Jon Hughes on Building A Quality Control Lab On A Budget (Aug 6, 2021).] | ||
| + | * [https://journals.asm.org/doi/pdf/10.1128/jmbe.00336-21 Bootleg Biology: a Semester-Long CURE Using Wild Yeast to Brew Beer.] | ||
==References== | ==References== | ||
Latest revision as of 17:15, 3 October 2024
Quality Assurance refers to the process if developing standard operating procedures for proactively avoiding quality problems [1]. In the brewing industry, this includes avoiding off-flavors from contamination, dissolved oxygen in beer, fermentation and ingredient issues, etc. Although quality assurance in the brewing industry covers a wide range of approaches such as verifying yeast health and pitching rates, recording fermentation data, performance tracking, etc. (see the External Resources below and Laboratory Techniques) [2], this article will focus primarily on avoiding microbial contamination through cleaning and disinfecting, particularly in the area of sharing a space for both pure culture fermentation and mixed fermentation beer.
Contents
Overview
Any microorganism that is introduced into a beer unintentionally and can survive in the beer is considered a "beer spoiler". Beer can be affected by contaminants via raw ingredients, poor sanitation, incorrect pasteurization, brewery environment air pollution, and inadequate ethanol production [3]. One survey of 38 craft beers in the Spanish market found that 68% of them had some presence of unwanted microbes, with beers under 5% ABV being more susceptible than higher ABV beers, indicating that craft breweries in particular may have a high degree of contamination issues [4]. While most microorganisms cannot survive in beer due to the hops, low pH, alcohol content, relatively high carbon dioxide, and shortage of nutrients, certain species are considered to be beer spoilage organisms due to their ability to adapt to brewing conditions (namely hops, ethanol, and low pH) and sometimes form biofilms that help them resist cleaning. Some are able to survive in beer and make a potential impact on the beer's flavor by producing acidity, phenols, turbidity/ropiness via exopolysaccharides (EPS), and/or super-attenuation (which can cause gushing or in extreme cases exploding bottles/cans) with just a few surviving cells. These effects can sometimes manifest days or even weeks after packaging, and longer storage or non-refrigerated storage can increase the potential for beer spoilers to negatively impact the beer. Bacteria species that have adapted to the brewing environment tend to be hop tolerant, but strains of the same species found outside of breweries are not tolerant of brewing conditions. It is thought that these species evolved to carry the genes to adapt to brewing conditions during the 5th to 9th centuries when hops were first being used in brewing, and that this evolution gave them a specialized adaption to the brewing environment where few competitors can survive [5]. Hop tolerant lactic acid bacteria have been found on the surfaces of many places in the brewing environment, including the fermentation area, bottling area, and cold storage. Hop tolerant lactic acid bacteria have been isolated from the air in at least one brewery in the fermentation and bottling areas [6].
Species of yeast and bacteria that are considered beer spoilers include Brettanomyces species, numerous Lactobacillus species, Pediococcus damnosus, Pectinatus species (anaerobe responsible for 20-30% of bacterial contaminations that produces acetic acid, propionic acid, acetoin, and 'rotten egg' like odors in contaminated beer), Megasphaera cerevisiae (7% of bacterial contaminations; inhibited below pH 4.1 and 2.8% ABV but can produce considerable amounts of butyric acid along with smaller amounts of acetic acid, caproic acid, isovaleric acid, acetoin, and hydrogen sulphide), Selenomonas lactifex, Zymophilus spp., diastatic strains of Saccharomyces cerevisiae, and some species from the Candida and Pichia genera. Most wild yeasts that can grow in beer in lab conditions are not considered largely impactful because of their limitatons to growing in the presence of ethanol or lack of oxygen, but they can become impactful on barrel aged beers where oxygen is present (Candida species, Pichia species, Torulaspora delbrueckii, Issatchenkia orientalis, Kluyveromyces marxianus, Debaryomyces hansenii, Zygosaccharomyces bailii, Zygosaccharomyces bisporus, Schizosaccharomyces pombe, and Kloeckera apiculata) [7]. Hop tolerant lactic acid bacteria make up the majority of contamination issues in breweries, with L. brevis making up more than half of the reported contaminations, and all lactic acid bacteria making up 60-90% of reported contaminations. A new species of Lactobacillus was recently identified called L. acetotolerans and was responsible for contaminating Goose Island's Bourbon County Stout, which is 60 IBU and 11% ABV. In sour beers with a pH below 4.3, only some lactic acid bacteria, Brettanomyces, and some wild Saccharomyces cerevisiae strains (which sometimes produce phenols, haze, over-attenuation, and/or over-carbonation) have the potential for unwanted growth, while beers with low alcohol, a small amount of hops, lower CO2 volumes (cask ales and beers dispensed with nitrogen, for example), and higher pH (4.4-4.6) are the most susceptible to contamination [8]. Pectinatus and Megasphaera are Gram-negative anaerobic species that produce a number of off-flavors in ales but not lagers (probably due to their preference for warmer temperatures). They are somewhat tolerant of hops (they can grow in beers with IBU's as high as 33-38 with one strain isolated from pickles reported to grow in beer up to 5% ABV and 80 IBU [9]) and often survive within the biofilms of other species in the brewing environment where the biofilm creates an anaerobic environment for them. They are sometimes found contaminating low ABV beers (under 5.2%) during packaging. They are not tolerant of pH below 4 and are killed at relatively low temperatures (58–60°C for one min) [5]. Zymomonas mobilis is a microaerophilic Gram-negative acetic acid bacteria that can withstand hops and can grow in bottled beer or casks where priming sugar is added and small amounts of air is present and produces high levels of acetaldehyde and hydrogen sulphide [8]. While the Gram-positive Staphylococcus xylosus bacteria, which grows on the skin of humans and animals, is not normally considered a beer contaminant, one strain was isolated from craft beer and was identified as the cause of increased turbidity, lactic acid, and succinic acid. It could grow at a pH between 3-7 (although only produced turbidity at a pH of 4-7), a temperature between 4–37°C, and as much as 8% ABV. This demonstrates that it is possible for species to adapt to living in beer other than the more typical beer spoilers [10].
Other species of microbes do not grow in beer but can become contaminants earlier on in the brewing process prior to primary fermentation (for example during kettle souring). These species include enterobacteria such as species of Clostridium, Obesumbacterium, Citrobacter, and Rahnella aquatilis, and wild Saccharomyces species that might not be able to grow in finished beer. These microbes produce dimethyl sulfide, organic acids, and 2,3-butanediol in high amounts which gives the beer an unpleasant rotting fruity or vegetal aroma [7]. Other species are considered "indicator" species because they do not directly cause spoilage of beer, but indicate that there is a hygiene problem. These include Acetobacter, Gluconobacter, Escherichia , and Klebsiella, as well as aerobic yeasts, all of which usually don't have an impact when present unless oxygen is also present. They can also produce slime that protects other microorganisms that can have a greater impact on the beer's stability [11][12]. Some species can contaminate yeast pitches. Pediococcus damnosus is frequently the cause of such contaminations and can cause diacetyl problems, as well as Pediococcus inopinatus, Pediococcus claussenii, L. casei, Selenomonas lacticifex, and Zymophilus raffinosivorans (although these are rarer to find in finished beer). Obesumbacterium proteus (which gives a parsnip-like smell and flavor) and Rahnella aquatilis can contaminate yeast pitches, and can inhibit fermentation and result in the beer finishing at a higher pH [8].
Biofilm forming spoilage organisms include a much wider range and higher frequency in beer tap systems than in brewhouses. This is due to the availability of oxygen and higher temperatures at certain points in the tap system, as well as poorer hygiene in tap systems as well as the difficulty to effectively clean plastic hoses. Of particular concern here is the ability of E. coli serotype O157:H7 to survive in tap systems, which has had a couple of documented occurrences in contaminated apple cider. Another study showed that aerobic yeasts were able to grow in dispensing lines, as well as L. brevis, and in many cases the draft lines were re-contaminated one week after cleaning, indicating that a contamination in draft lines is difficult to remove [13][11].
Sources for contamination in breweries can occur as "primary" contaminations (yeast pitching, and brewhouse related contaminations), or as "secondary" contaminations (packaging and cellaring), as well as in tap systems. They are usually not sudden occurrences, but a result of the continued growth of microorganisms in a problem area. Historically, re-pitching yeast was often a source of contamination; however, more recently this has become less of a source for contaminations due to better education and techniques. Typical sources for contamination also include unclean equipment such as thermometers, manometers, valves, dead ends, gas pipes, leaks in any part of the system (especially at heat exchangers), wort aeration equipment, and even worn floor surfaces. More than half of the documented contaminations come from the packaging system. These are typically the sealer (35%), the filler (25%), the bottle inspector (10%), dripping water from the bottle washer (10%), and the environment close to the filler and sealer (10%). In regards to the environment as a source of contamination, this has been found to be from airborne contaminants near the filler and crowner in open bottles on their way from the bottle washer to the filler and from the filler to the capper. The higher the humidity and the more airflow, the more chances of airborne contamination [13][8]. For example, Pectinatus, while mostly found in lubrication oils, water systems, floors, water condensed on ceilings, etc., it can also survive on aerosols in the air and is thought to possibly transferred to beer that is being packaged via the air [5]. In tap systems at taverns, 'one-way' valves that are attached to kegs have been found to be a source of contamination, as well as the dispensing line [13][8].
Thompson et al. (2024) divided the brewing environment into three "zones": areas that have direct contact with beer, such as the brewhouse, areas that are adjacent to beer production areas that have no physical barrier from the beer production area, and areas that have a physical barrier between the beer production areas, such as offices, tap rooms, etc. They took samples from all areas in three small microbreweries. The study found that racking arm valves were one of the worst offenders of housing contaminating microbes, although the results were skewed by one of the breweries having a higher load of contaminants in general between the three microbreweries. Proper cleaning, sanitation, and maintenance of racking arms is recommended to reduce biofilm formation. Other areas that harbored bacteria included the bottom valve, carb stone valve, spray ball valve, and the yeast pitch. All areas of the brewery that were swabbed show at least some bacterial growth, demonstrating that all areas of the brewery are potential sources of contamination. This study noticed that one brewery in particular had a higher occurrence of spoilage microbes; the study speculated that this might be due to cleaning and sanitation protocols during packaging, and heavy loads of contaminants were found in the packaging system [14].
See also:
- The Barrel wiki page.
- Diagram overview of bacterial and fungal species reported at all stages of beer production.
Microbe Populations in Mixed Fermentation Breweries
Bokulich et al. (2015) studied the microbial populations throughout a brewery (presumably Allagash) that produces clean beer, mixed fermentation sour beer, and spontaneously fermented coolship ale. They found that most of the microbes living in breweries were introduced from the ingredients such as malted barley and hops, and many populations were confined to specific rooms or areas within the brewery. Some species did spread to other rooms, presumably through human and insect vectors. Beer resistant lactic acid bacteria spread throughout the brewery (although more abundantly found near packaging equipment and fermenters that were filled with sour beer), but the clean beer was largely uncontaminated. Physical partitions and walls appeared to help inhibit the spread of microbes from room to room [15].
Ale yeast was found throughout the brewery, especially in the fermentation cellar. Malted grains were determined to be the highest source of potentially contaminating microbes (mostly Pediococcus, although the PCR method used in the study was not adequate for detecting microbes on grain) in the hotside areas of the brewery compared to other microbe sources such as human skin, outdoor air, soil, saliva, feces, water from the plumbing, etc [15]. Other studies have identified grain dust and aerosols as airborne vectors that disperse a range of lactic acid bacteria [7]. Hops were determined to be the highest source of microbes in the cellar fermentation areas (Pediococcus spp, Lactobacillus lindneri, and L. brevis were detected on pellet hops, although the presence of hop tolerant genes was not detected from microbes found on hops), and yeast was the highest population in fermenter and packaging areas. While human skin was a minor contributor to microbe populations found in the brewery, oak barrel surfaces were populated with microbes from unknown sources. Therefore, it was determined in this study that raw materials are the major source for potential contaminants in a brewery, although minor sources such as sinks are still potential problems and hop tolerant species were linked to the purposeful inoculation of souring microbes in the sour beer production itself [15]. L. lindneri is highly hop tolerant, difficult to detect on MRS media, and has been found to survive suboptimal heat pasteurization temperatures. L. brevis strains that have been growing in beer are also hop tolerant, although some strains have been found to lose their ability to grow in beer if they are grown in lab media after a few generations and strains not found in breweries are not hop tolerant, which suggests that strains found in beer have adapted to that environment. Both of these species also have a smaller cell size when they have been growing in beer, and can pass through microbial filtration systems [5].
Seasonality played a minor role in the populations of microbes throughout the brewery, with ale yeast and Candida santamariae spreading from the fermentation and packaging area to the rest of the brewery from fall to summer (it was proposed that the warming from fall to summer played a role in the spread of ale yeast and Candida santamariae throughout the brewery). Micrococcus and Kocuria were found in more localized areas such as the floors and other surfaces in the barrel room, cellar, and packaging room. Acetobacter and Lactobacillus were found specifically in areas where a lot of wort or beer was being processed (conveyor belts and floors below the packaging equipment, hotside and cellar area sinks, and sample ports on kegs and fermenters). Lactobacillus was more common on surfaces where sour beer production was (fermenters and barrel surfaces), with floor surfaces having a more diverse mixture of LAB species [15].
Microbes with hop tolerant genes were found more abundantly in the fermenter and packaging areas (filler heads, below the bottling line belt, packaging sink, and a keg faucet) compared to microbes found on pellet hops (which were determined to not be a source of contamination), kegs, or barrel bungs, and was associated with where beer was being processed, particularly mixed fermentation and spontaneously fermented sour beer [15].
In the case of mixed cultures or contaminations that contain Pediococcus damnosus., Lactobacillus brevis, or Lactobacillus lindneri, and possibly other species of these two genera, it is possible for the cells of these bacteria to adhere to the cells of brewers yeast. This can cause the yeast to prematurely drop out of suspension during fermentation, resulting in under attenuated beer. This function of the bacteria is thought to contribute to the slower development of the bacteria after the yeast has dropped out of solution. Other species of Lactobacillus such as L. casei/paracasei, L. coryniformis and L. plantarum as well as species of Leuconostoc are very intolerant of hops and are therefore only considered a threat against beer that is very lowly hopped [5].
Biofilms
Many microorganisms can form biofilms which is defined as a community of cells of one or more species that are attached to each other and/or a surface and are embedded in a matrix of extracellular polymeric substances (EPS). EPS consists of polysaccharides and proteins that are produced by the microorganisms and expelled out of the cells, similar to a pellicle. Biofilms allow microbes to survive less vigorous cleaning and sanitizing regiments and chemicals and has become a concern in the food industry as well as in the brewing and winemaking industries [16]. Biofilms most often form in the packaging system somewhere, but can also be found on side rails, wearstrips, conveyor tracks, drip pans, and in-between chain links [13].
Bacteria and wild yeast form a biofilm in two stages, which are determined by a number of variables. In the first stage, the microbes remain in their "planktonic" form (floating around in the liquid), but they begin to adhere on surfaces and to each other as those surfaces. Other species of microbes can also be adhered to during this phase. The second stage is where the microbes start producing exopolysaccharides (EPS) which helps them bind together in a matrix, along with any available proteins and exopolymers produced by the bacteria. A large portion of biofilms is actually water (80-80%) as this allows the microbes to remove waste and consume nutrients. This matrix helps the microbes resist antibiotics, UV radiation, and cleaning chemicals. Gene exchange also occurs more frequently. At the end of this second stage, the microbes become attached to surfaces in such a way that is permanent without the use of cleaning chemicals. This is known as the microbe's "sessile" form (immobile). Bacteria in this form continue to multiply, and upon maturation of the biofilm, eventually, planktonic cells begin to be produced and released from the biofilm to find new homes. They also display different phenotypes, which might contribute to their ability to resist cleaning chemicals. Rough surfaces, scratched surfaces, jagged edges, and pores are more prone to biofilm formation due to the higher surface area. Hydrophobic surfaces, such as Teflon and other plastics, are more prone to biofilm formation than hydrophilic surfaces (glass and stainless steel). Nitrile butyl rubber (NBR) was found to inhibit biofilm formation when new, but as the material breaks down biofilms are able to grow [17]. Biofilm formation is strain specific rather than species specific; some strains can form thicker biofilms than others within the same species and faster, and some strains of lactic acid species are not good biofilm producers [11]. Wild strains of S. cerevisiae that carry the "Flo11p" gene tend to form biofilms which suggests that this ability is important for survival in the wild, but domesticated strains have mostly lost this ability probably due to evolving under nutrient-rich environments (human-controlled fermentation), and their planktonic form may give them an advantage in nutrient-rich liquids, especially during spontaneous fermentation where their ability to be mobile might help them compete against other species of microbes [18][19]. Full biofilms can form within 2-4 days for some strains, while 10 days is required for significant biofilm formation in other strains. For example, one strain of Lactobacillus brevis isolated from draft beer did not form any biofilm, while another strain of L. brevis tested was a strong biofilm producer. Similar results were observed for Brettanomyces strains. In general, mixed cultures form stronger biofilms than single cultures. The presence of soil (biological residue) encourages biofilm formation [11]. The presence of sweeteners or sugar also encourages the formation of biofilms. In one study (Storgårds 2006), biofilm forming species were found to begin attaching themselves to brand new sterile stainless steel surfaces within 2-12 hours after the new equipment was used for production [20].
The efficacy of different chemicals to kill microbes within a biofilm isn't widely studied in the brewing or wine industries, partly because testing procedures are laborious and difficult to standardize. Studies have found that alcohol-based disinfectants (ethanol and isopropyl alcohol) and hydrogen peroxide-based disinfectants were effective at killing microbes within a biofilm, and peracetic acid disinfectants were not as effective. A higher concentration of peracetic acid (from 0.25% to 1% of products containing 4-15%) was required to be more effective than lower concentrations. However, these disinfectants did not kill all of the cells without a cleaning regiment first. Yeast biofilms, in general, are more susceptible to cleaning chemicals than bacteria biofilms. Biofilms that are formed under static conditions (still or dried up liquid) are more resistant to disinfectants than biofilms that form under flow conditions (movement of liquid) [11][21].
See also:
Spores
Some species of fungi and bacteria can form spores. Yeast generally forms spores in order to reproduce sexually. Their sporulated forms are not a mode of protection from disinfectants and are therefore killed by normal sanitation methods. Bacteria and molds form spores as a mode of survival. For example, some dangerous types Clostridium botulinum spores require 250°F (121°C) for 3 minutes to be killed, which is the requirement for canned goods [22][23][24]. Spore-forming species of bacteria, however, are not considered beer spoilers [25]. Thus, the challenge of killing yeast or bacteria spores is irrelevant in most beer and wine production. There are some extraneous brewing methods where bacteria spores should be considered, for example wild yeast isolation safety, mold formation during fruit fermentation or barrel aging, and the long storage of unfermented wort. Lactobacillus species do not form spores.
Hygienic Methods For Avoiding Contamination
General Approaches
- Clean and disinfect all equipment that comes into contact with the beer or wort, including beer dispensing systems, and keep them in prime condition.
- Use the maximum concentrations, exposure times, and hottest temperatures (considering temperature limitations of plastics and glass) suggested by the manufacturers of any given cleaning/disinfectant product.
- Clean first using an effective cleaner, and then apply a disinfectant/sanitizer as a second step.
- Use 180°F (82°C) hot water for 60 minutes to disinfect stainless steel and other heat tolerant materials (check with your manufacturer to make sure that the vessel is rated to withstand fast hot/cold cycles; vacuum or pressure relief valves should be used in order to avoid imploding due to fast temperature shifts for some equipment).
- For any plastics that cannot be treated with heat, especially tubing, keep separate plastics for use with potential contaminants such as Lactobacillus, Pediococcus, Brettanomyces, and diastatic strains of Saccharomyces cerevisiae.
- Replace rubber and plastic parts such as gaskets as often as recommended by the manufacturer or when wear is apparent.
- When operating a commercial brewery, invest in a quality control lab and procedures to identify inefficient hygiene practices.
- Use a separate packaging system for sour beer unless the packaging system can be sanitized with hot water or caustic (foam disinfectants that are often used in packaging lines have been reported to be not as effective against removing biofilms).
- The more surface area that equipment has, the more prone it is to biofilm formation. Horizontal surfaces are more prone than vertical surfaces to biofilm formation.
- Heat pasteurize, and store beer and yeast at low temperatures. Beer filtration and pasteurization are effective ways to reduce the chance of contamination.
- Non-alcoholic beer and beer under 1.3% ABV grew bacteria 2-5 times more than 4.5% ABV lager beer in one study, and the authors concluded that long-draw draft lines should not be used to serve these types of beers [26].
Reducing Microorganisms
Several generalized procedures are used for limiting the number of unwanted microorganisms. These include acid washing yeast that is re-pitched (kills bacteria but not wild yeast that is tolerant of low pH), keeping beer cool (slows the growth of microbes in general), filtration (removes yeast), pasteurization (kills vegetative cells in the finished beer, but not spores - most beer spoilers are killed at 15 pasteurization units (PU) and all are killed at 30 PU using a recommended pasteurization temperature of 66°C ), and aseptic or hygienic packaging. Note that some strains of Lactobacillus have been shown to survive pasteurization temperatures; see Lactobacillus Heat Tolerance. Packaging systems should be frequently flooded with hot water between 80-95°C or saturated steam (every 2 hours in the summer and every 4 hours in the winter). UV light or disinfecting chemicals are also used. The filler and crowner should be disinfected frequently as well. Packaging in an aseptic room with HEPA filtration and higher air pressure within the room compared to outside, along with special clothing, is another method that larger breweries use to remain aseptic [13].
Most brewing equipment should be designed for good hygiene. Pits and crevices should be avoided, and all surfaces should be smooth when possible. All equipment and pipelines should be self-draining. Valves are a typical source of contamination because they are not easily CIP'ed, especially plug valves and ball valves (although butterfly, gate, and globe valves are also difficult to CIP) [13]. Horizontal surfaces and wet surfaces are more prone to biofilm formation. In one study that compared biofilm formation in bottling lines versus canning lines, it was found that canning lines develop less microbial biofilms and contaminations than bottling lines due to not having rinsing stations, labeling stations, and simpler constructions than the bottling lines that were studied [20]. However, some canning lines cannot use caustic for cleaning, or it is not common practice, but use foaming agents instead which are less effective at removing biofilms (see efficacy of cleaning agents below). The lack of use of caustic cleaners in canning lines has been identified as a source of contamination issues with diastatic strains of Saccharomyces cerevisiae in canning lines [27].
Some other methods have been proposed by scientists as being novel ways to reduce unwanted microorganisms. These include exploiting naturally produced toxins. For example, some lactic acid bacteria produce bacteriocins which can kill other bacteria. Some strains of wine yeast can produce zymocins that kill other species of yeast. Such methods are viewed as being fairly extreme. Advances in genetic engineering techniques make these approaches technically possible, however, there currently exists a commercial stigma against genetic modification. Additionally, there are many types of toxins which target only specific species, so anticipating which species should be targeted could be challenging [8].
Yeast Washing
Yeast washing is the practice of exposing a yeast slurry to extreme acidic conditions in order to destroy bacteria contaminants in the slurry, and it has been used for over a century in the brewing industry to help reduce the potential for lactic acid bacteria spoilage. Although techniques might vary throughout the brewing industry, the most typical technique is to add phosphoric acid to a slurry of yeast until a pH of 2 is reached, and then the slurry is stored for 2 hours at 5°C (41°F). While phosphoric acid is a good choice for acid washing because of being inexpensive compared to other acids, its tendency to not kill yeast, and its lack of affecting beer flavor, it also does not kill some contaminants such as Shimwellia pseudoproteus. It has also been proposed that chlorine dioxide, a disinfectant that is often used in the vegetable, meat, and water treatment industries, can be successfully used to wash a yeast slurry, with the first study on this reporting that a concentration of 78 mg/L (concentration value is for the entire slurry) and stored for 30 minutes at 8°C (46.4°F) was effective [28].
Lysozyme, an enzyme that is often extracted from hen egg whites, is known to inhibit Gram-negative bacteria such as Lactobacillus but not Gram-positive bacteria such as Acetobacter, and has been shown to be an enzyme that can help inhibit spoilage bacteria in wine and cider fermentations [29]. Lysozyme is normally added to wine with a stuck fermentation or to limit malolactic fermentation, and several yeast companies offer a lysozyme-based product [30][31]. It has also been suggested to be useful for limiting lactic acid bacteria in yeast slurries, but one experiment reported that the sensitivity of different species of lactic acid bacteria varies, with Pediococcus inopinatus, Lactobacillus brevis, Lactobacillus brevisimilis showing similar levels of sensitivity, but L. linderi showing less sensitivity. Bacteria were inhibited more at 22°C than at 4°C. At 300 mg/L, although lactic acid bacteria was inhibited, it was not killed completely [32]. Nisin has also been proposed as a potential preservative that can be added to wort during boiling or cooling as well as to the fermenter in order to limit the growth of lactic acid bacteria by up to 90% [33].
The homebrew practice of mixing distilled or sanitized water into a yeast slurry, letting the slurry settle into three layers, and then removing the bottom and top layer and re-pitching or saving the middle layer, is different than "yeast washing". This process is known as "yeast rinsing", and is primarily employed by homebrewers who wish to separate trub material from their yeast slurries before reusing the yeast slurry. This might have the benefit of removing unwanted flavors from the slurry (although there is a lack of evidence that we know of for this claim [34]) or hop material that could inhibit yeast growth, but it does not inhibit lactic acid bacteria or any other contaminants (in fact, this process increases the chances of contaminating the yeast slurry). See this AHA article for more details on yeast rinsing.
Cleaning and Sanitizing
The goal of cleaning is to remove as much biomaterial as possible, while the goal of sanitizing is to reduce the population of viable microbes as much as possible and prevent them from growing on surfaces during the non-production time. It's been shown that chemical cleaners are better at removing biofilms than sanitizers and disinfectants, and sanitizers that kill cells in suspension may not be effective at killing cells within biofilms. Complete removal of unwanted microbes within biofilms can be achieved by first using a cleaning agent to remove the biomass followed by a sanitizing/disinfecting agent. CIP procedures may not be enough to remove biofilms without high turbulent flow with spray nozzles and the use of heat (low cleaning temperatures are not effective at removing biofilms). Chlorinated alkaline detergents were found to be the most effective at removing biofilms [11]. Below is a typical CIP process according to Erna Storgårds (2000); CIP processes at room temperatures are not adequate enough to remove biofilms, so use hot temperatures when applicable. Use the highest chemical concentrations recommended by the vendor. Also, the higher the velocity of the cleaning fluid through the system, the more efficient it is at removing biofilms:
| Action [11] | Temperature | Duration |
|---|---|---|
| Pre-rinse | cold or hot | 5-10 min |
| Alkali cleaning with 1.5-4% sodium hydroxide (caustic) | cold or hot (60-85°C) | 10-60 min |
| Rinse | cold | 10-30 min |
| Acid wash (phosphoric, nitric, or sulphuric acid) | cold | 10-30 min |
| Rinse | cold | 10-30 min |
| Disinfection (chemical such as peracetic acid or hot water at 85-90°C) | cold (or hot if using just water) | 10-30 min with chemical, or 45-60 min with hot water |
| Rinse (might contain a low concentration disinfectant) | cold | 5-10 min |
Open surfaces such as bottle inspectors, fillers, and conveyor belts in the packaging line should be first rinsed with water, then cleaned with a foaming agent, rinsed again with water, and then sprayed with a disinfectant solution and a final rinse. Components that cannot be visually inspected should be dismantled and inspected. Rubber gaskets and sealings have been found to house biofilms, especially after deteriorating, and so they should be inspected and replaced as needed. NBR rubber has been found to inhibit biofilms when new, and EPDM rubber has been found to be anti-bacterial towards some bacteria [11].
See also:
- MBAA Podcast Episode 253 CIP Fundamentals.
- Barrel Sanitizing.
- Joe Idoni's heat sanitation based SOP.
- MTF thread on brewing mixed fermentation beers with clean beers in a commercial brewery.
- Another MTF thread on sanitation.
- MTF thread on using one keg cleaner to clean regular beers and sour beers in commercial breweries.
- "A Clean Fight: The Science of Hygienic Brewing" by Colin Kaminski in Brew Your Own Magazine.
- Brian Hall demonstrates steam sanitizing a corny keg.
- "Assessing Contamination Risks of Non-Conventional Yeasts in Breweries," bachelors thesis for Ronja Eerikäinen (yeast study only); see also an interview with Eerikäinen on the The Brü Lab podcast episode 16.
- "Reducing Dissolved Oxygen: Acid and Detergent Cleaning of Brite Tanks," white paper by Dana Johnson Technical Director, Craft Brewing, Birko. As appeared in The New Brewer, July/August 2011.
Oak Barrels
See the Barrel wiki page.
Pasteurization
Pasteurization is measured in terms of "pasteurization units" (PU). One PU is equal to exposure of 60°C (140°F) for 1 minute. The total PU is determined by plotting time against temperature in degrees Fahrenheit. A total of 15 PU's has been given as the target for pasteurizing beer. The following equation can be used to calculate PU's using different temperatures and times [35]:
There are two types of pasteurization methods used in brewing: tunnel pasteurization and flash pasteurization. In tunnel pasteurization, which is more widely used in breweries, cans or bottles of packaged beer is moved slowly through a tunnel of fixed temperatures. In flash pasteurization (or plate pasteurization), large quantities of beer are pasteurized at the same time via a heat exchanger and is usually performed before the beer is packaged [8]. Since thermal death rates for beer spoilage organisms has been identified to be 140°F (60°C) for 15 minutes [35][36], this is the baseline temperature and time for pasteurization, although higher temperatures and shorter times are used for some pasteurization methods (see the below links). The complete thermal death of Brettanomyces in wines has been reported to be 50°C for 5 minutes. [37][38]. Pediococcus is generally not tolerant of temperatures over 45°C (see Pediococcus). Some strains of Lactobacillus have been shown to potentially survive pasteurization temperatures for at least some amount of time; see Lactobacillus Heat Tolerance for more information.
Microfiltration is an alternative technology to heat pasteurization that can be used to pasteurize beer. Microfiltration uses a set of membranes, usually in the 0.45–0.65 μm range, for filtering bacteria and yeast. Bacteria have a cell size of about 5-10 μm and yeast species have a cell size of about 5–16 μm, while flavor compounds such as phenols are filtered out when using a smaller diameter filter such as 0.2 μm. One study by Bernardi et al. (2019) found that filtration with polyethermide membranes removed around 1-2 IBU, ~30% of yeast-produced phenolic compounds (most polyphenols from hops were not filtered out), and larger tannins (which were only a small portion of the total polyphenol content). The antioxidant activity was largely not impacted. After filtration, the beers were 26%-33% lighter in color, depending on the style of the beer, and were 100% clearer. The filtration that was used, which was 1.2 μm, also produced fully pasteurized beers [39].
Diastatic strains of Saccharomyces cerevisiae can have a wide range of temperature tolerance. Strains that can form ascospores or vegetative cells can be more heat tolerant. One thesis paper reported it taking 9 minutes at 60°C to kill 90% of a strain of diastatic S. cerevisiae in ascospore form [40].
See also:
- Barrel Sanitizing.
- "Pasteurization in Beer Production" and "Pasteurization in Breweries"; AEE - Institut für Nachhaltige Technologien wiki.
- Heat Treatments and Pasteurization standards for milk processing.
- "Is flash pasteurization right for your craft beer?" by Chris Crowell in Craft Brewing Business website (details case studies for temperatures and times).
- "Understanding the Risk of Can Pressure Failures" interview with Jim Kuhr on MBAA Podcast episode #240.
- MBAA Podcast "Pasteurization At Goose Island".
- Example homebrew method for heat pasteurization by Pappers_ on HomebrewTalk (do not attempt this with highly carbonated beverages; bottles will break).
- MTF thread on using sulfites and sorbate to stabilize fermentation in beer.
- "Keeving," a traditional method of achieving carbonation in sweet ciders (not pasteurization, but potentially a related application).
Draught Line Cleaning
See the Brewers Association Draught Beer Quality Manual.
Efficacy of Cleaning Agents
Commercial cleaners and disinfectants

Sodium hydroxide (caustic), EDTA (ethylene diaminetetra-acetic acid), chlorinated disinfectants, and hydrogen peroxide-based disinfectants such as Pur-Ox from Birko or Lerasept-O from Loeffler are effective at breaking up biofilms when used in their highest recommended concentrations [11][41][42]. Foaming agents that are often used in packaging lines for cleaning, however, might not be as effective. One study found that one foaming agent (VK10 Shureclean, which is sodium alkylbenzenesulphonate) required two times the maximum concentration that is recommended by the manufacturer to completely remove biofilms. In comparison, all of the sodium hydroxide (caustic) based cleaners that were tested were effective at completely removing biofilms in concentrations that were below the vendors' recommended maximum concentrations [43]. Another study tested six different types of cleaners were tested to see how well they removed the biofilms: keytones + surfactant detergent, quaternary ammonia + surfactant detergent, sodium hydroxide (caustic soda), sodium carbonate (soda ash), sodium hydroxide + surfactant (alkaline detergent), and chlorine (sanitizer, not a detergent). They found that only caustic soda was consistently efficient at removing the biofilm (the caustic, ammonia + surfactant cleaner, and the quaternary ammonia + surfactant detergent all efficiently removed cells that were adhered to a surface over 6 hours and were determined to be good for stopping biofilm formation from occurring in the first place) [44]. Peracetic acid (PAA) has also been shown to be effective against biofilms in the highest recommended concentrations but isn't as effective as the previously mentioned cleaners and should be used after a caustic cleaning cycle [45][46], but its effectiveness decreases below 20°C. Chlorine and iodine-based disinfectants destroy microbe at colder temperatures, however, they are less effective in the presence of wort or other residues. Chlorine-based disinfectants can cause pitting in stainless steel if left in contact for too long, and some stainless steel manufacturers recommend not using chlorine-based disinfectants at all (refer to your equipment and chemical manufacturers). Hot water is one of the most effective disinfectants, however, dry heat is not as effective at killing bacteria (one strain of L. brevis was able to withstand 80°C dry heat for 60 minutes) [11]. Dry heat at higher temperatures will sterilize at 170°C for 1 hour or 190°C for 12 minutes and can be used to sterilize many metal and glass instruments. Flaming surfaces kills within seconds [47].
Ethanol and isopropyl alcohol have also been found to be very effective sanitizers. The downside to these sanitizers is that they are flammable and are very volatile, evaporating quickly after use. One experiment by Elliot Parcells & Josh Pohlmann from Bells Brewery claimed that iodophor was an ineffective sanitizer at concentrations of 25 ppm, however, their results have been questioned due to their methodology [48].
Sulfur dioxide has been used as a sanitizer in some cases in winemaking; see this MTF thread for more information. SO2 is also used as a microbial stabilizer in finished wine. 50 ppm of total SO2 can inhibit the growth of lactic acid bacteria in finished wine [49]. 2.1 mg/L of molecular SO2 is needed to inhibit Brettanomyces in finished wine [50]. See this article from Cornell on an explanation between total SO2, free SO2, and bound SO2.
Caustic cleaners can errode glass when the pH gets above 9.0 [51][52].
Enzymes have been reported as a solution to helping to clean biofilms from within dispensing tubing. Dispensing line cleaners have been reported to be inneffective at cleaning established biofilms due to the protective exopolysaccharide (EPS) layer of biofilms and the proteins that yeast cells excrete during flocculation, which help them adhere to each other and to surfaces. Polysaccharases, polysaccharide lyases, and to a lesser extent, proteases, will catalyze EPS and proteins. Microbes secrete these enzymes themselves in order to free themselves from the biofilm when their environment becomes stressful, and the effect of a particular enzyme degrading a particular EPS depends on other EPS types in the biofilm; degrading one EPS type can lead to the growth of another species present in the biofilm. Thus, the effectiveness of enzyme cleaning depends on the enzymes used and the types of microbes in the biofilm. This is one potential problem with using enzymes for cleaning [53].
For example, Walker et al. (2007) found that a mix of the enzymes alpha-amylase, beta-glucuronidase, glycose oxidase, dextrinase, protease, and pectinase followed by the use of a line cleaning agent reduced the number of viable S. cerevisiae, Lactobacillus brevis, and Leuconostoc mesenteroides by 2 logs, but the number of Flavimonas oryzihabitans increased. A doubling of the enzyme content resulted in more effectively elimating Flavimonas oryzihabitans, but it still wasn't as effective as elimating the other microorganisms. Additionally, the enzymes were less effective on dispensing lines than they were on stainless steel. On dispensing lines, the bacteria were reduced, but yeast was not. The study concluded that a broad range of enzymes should be used, and might assist with breaking up biofilms when used in conjunction with line cleaning agents, but this depends on the types of microbes/EPS in the biofilm and some microbes might be more resistant to this treatment than others [53].
Homebrew cleaners and disinfectants
Five Star Star San
Five Star Chemicals product Star San is a popular acid anionic sanitizer sold to homebrewers because of its relative safety and ease of use. Claims that acid anionic sanitizers are not effective at killing yeast have been made on various internet forums [54][55][56]. These claims are cited in various food science pamphlets, blogs, and websites, and appear to be based on the food science textbooks, "Principles of Food Sanitation," by Norman G. Marriott and Robert B. Gravani (2006) and "Basic Food Microbiology" by George Banward (1989), which contain conflicting information about the effectiveness of acid anionic sanitizers. Neither of these textbooks contain experimental data nor references to experimental data. The statements in these books conflict with each other. Marriott and Gravani claim, "(Acid anionic sanitizers) have limited and varied antimicrobial activity (including poor yeast and mold activity)... The antimicrobial effect of acid anionics appears to be through reaction of the surfactant, with positively charged bacteria by ionic attraction to penetrate cell walls and disrupt cellular function." Banward's claims of acid anionic sanitizers are, "Advantages: Active against a wide spectrum of microorganisms including thermodurics, controis phage and most yeast strains. Disadvantages: Slow activity against sporeformers, not effective in destruction of most spores." Furthermore, the provided explanation, which is that acid anionic sanitizers supposedly don't work effectively against yeast and molds is because acid anionic sanitizers are negatively charged and yeast are also negatively charged yet bacteria is killed because it is positively charged, is biologically incorrect (this masters thesis appears to be the source of this incorrect information). According to Dr. Bryan Heit of Sui Generis blog, both yeast and bacteria have negatively charged cell walls, and this fact has been well established in microbiology since the 1940's (Dr. Heit has published several peer-reviewed scientific studies on cell wall polarity).
Star San has only been officially tested by Five Star Chemicals against the pathogenic bacteria species E. coli and S. Aureus, which is the minimum baseline required by the EPA to be labeled a "sanitizer" [57]. While we are not aware of any publicly available published studies on the efficacy of StarSan to kill yeast, several studies with other acid anionic sanitizers have confirmed that they are effective against yeast. Lee et al. (2007) found that an acid sanitizer very similar to Star San that uses citric acid instead of phosphate but the same surfactant (sodium dodecylbenzene sulfonate) took 5 minutes to kill Saccharomyces cerevisiae, E. coli, and Listeria innocua at room temperature (some species were killed faster than others with the E. coli actually being more resistant than the yeast), and one minute if the sanitizer was heated to 40°C on both metal and LDPE plastic (they compared the acid anionic sanitizer to 35% hydrogen peroxide, which killed all organisms with 15 seconds, indicating that this acid anionic sanitizer is effective at killing yeast, but it takes longer than a stronger chemical such as hydrogen peroxide). This study did not make mention of biofilms, however, the cultures were allowed to grow and dry overnight which could have allowed for biofilm formation [58]. Five Star Chemicals also recommends 5 minutes of contact time with Star San. Winniczuk et al. (1997) found that three phosphoric acid anionic sanitizers ("CS-100" and "CS-101-lf" by Chemical Systems of Florida, and "Clear-Clean" by Pelican Brand) were less effective at killing yeast than bacteria in the timeframe tested (1 minute contact time), but they were still effective at killing yeast at high concentrations (peracetic acid also required a higher concentration to kill yeast than bacteria). However, one of the acid anionic sanitizers tested was more effective than the other two, indicating that the chemical makeup of the particular acid anionic sanitizer has an impact on how effective it is as a sanitizer relative to other acid anionic sanitizers. Additionally, they found that peracetic acid, iodophor, and chlorine dioxide required less concentration than the acid anionic sanitizers to be effective (again, tested at 1 minute exposure time) [59].
See this MTF thread for a more extensive explanation of why skepticism should be applied to the claim that acid anionic sanitizers are not effective at killing yeast.
Tips for using Star San:
- Completely remove all soils as soon as possible from equipment after use using an effective cleaning agent.
- Apply Star San before using the already cleaned equipment.
- Dilute the Star San in distilled or reverse osmosis water so that the pH is not buffered by chemicals in tap water.
- Leave in contact with surfaces for 5 minutes or more.
- Optional: warm the Star San and water solution to 40°C/104°F.
Iodophor
A popular alternative to Star San is iodophor, which is an iodine-based sanitizer.
Elliot Parcells & Josh Pohlmann from Bells Brewery tested the efficiency of various sanitizers to kill Lactobacillus, Brettanomyces, and diastatic strains of Saccharomyces cerevisiae', and claimed that iodophor was ineffective at concentrations of 25 ppm, which is the maximum concentration of iodophor for it to be considered a no-rinse sanitizer. The methodology of this experiment has received some criticism that might invalidate its results. They diluted yeast/bacteria cultures on a 1:10 ratio into iodophor, thus diluting the iodophor below its recommended dilution and exposing it to concentrations of microbes far beyond what it was designed to kill. Testing the ability to sterilize a bulk solution does not translate to the ability of the sanitizer to sanitize trace surface microbes after proper cleaning [60].
Five Star PBW
PBW is a replacement for caustic and is much safer to use and dispose of. It is non-corrosive and safe to use on stainless steel and plastics. We are not aware of any studies that examine the efficacy of P.B.W. to remove biofilms, nor any other percarbonate-based [61] cleaners efficacy in a brewing environment.
Tips for using PBW on equipment that may have had biofilm formation [62]:
- For circulated cleaning, use 1 to 3 ounces per gallon depending on the soil load, and heat to 180°F. Recirculate for 30 minutes (if using this product on plastics, heat the water that is mixed with the PBW to be as hot as the manufacturer of the plastics recommends, and extend the circulation time to an hour or two).
- For non-circulated, soaking, use 6 to 8 ounces per gallon of 180°F water (if using this product on plastics, heat the water that is mixed with the PBW to be as hot as the manufacturer of the plastics recommends), and soak overnight. Note that there have been reports of high concentrations of PBW breaking down PET bottles over time when soaking [63]. For example, BetterBottle™ recommends using 5 grams per liter of 125°F water (0.67 ounces per gallon, or one tablespoon per gallon) of PBW and cleaned with agitation/circulation instead of soaking. They also recommend using an enzymatic cleaner such as Seventh Generation Free and Clear Natural 2X or Super Pro-zyme Enzymatic Cleaner instead of PBW [64].
- Rinse thoroughly with hot water (cold water may not remove some of the residual chemicals).
Alternatives to PBW include:
See Also
- "A Complete Guide to Cleaning and Sanitation" by John Palmer.
- "How To Brew". Chapter 2 Cleaning & Sanitizing, by John Palmer, 4th Edition, 2017.
- BetterBottle Guide to cleaning and sanitizing PET.
Quality Control
(To do)
Quality Control
Quality Control is the process of identifying quality problems in the product, and is a reactive process aimed at correcting a detected problem [1].
- ATP bioluminescence system monitors for contaminants. This method uses light to detect organic matter, but cannot identify the microorganisms that are present. It also indicates whether the biological matter is low enough to employ disinfectant.
- Swabbing and cultivating on agar and PCR.
- Direct epifluorescence filter technique (DEFT)/microcolony method.
- Direct impedimetry.
- Flow cytometry.
- DGGE/TGGE (separation of heterologous sequences by chemical/heat denaturation).
- Clone Libraries (Sanger sequencing of transformed E. coli clones containing diagnostic gene amplicons).
- qPCR
- Illumina Sequencing
- 454 Life Sciences Pyrosequencing
- PCR DNA dipstick for rapid detection at different stages of the brewing process.
- Disposable electrochemical biosensors for Brettanomyces bruxellensis in wine [65]
- PDF from Richard Preiss with a summary mostly derived from Jorgenson, E. (2017). An Overview of Bacteria Found in Brewing Ecosystems. Master Brew. Assoc.
- RNA-FISH (Fluorescense In Situ Hybridization) technique for detecting Brettanomyces contamination should use a red-emitting fluorophore.
- Surface enhanced Raman spectroscopy phylogenetictree for genosensing of Brettanomyces bruxellensis yeast on nanostructured ultrafine glass supports.
Beer adapted strains of Brettanomyces and lactic acid bacteria have been found to be more difficult to culture on agar than when they are not adapted to the environment [5]. Malt agar has been shown to be more effective at showing growth than some other types of agar (Dekkera Medium, Universal Beer Agar, Potato Dextrose Agar, and Beer Agar). YPD with 10 ppm Cycloheximide was not tested in the study, but Nick Impellitteri, the owner of The Yeast Bay, reports that this media works well for him [66], and DBDM media has also been reported to work well for growing Brettanomyces. In addition, it is recommended that 100 mL samples are taken since beer adapted contaminants are harder to grow on media, and at least 7 days of incubation time should be allowed for Brettanomyces to show signs of growth [67] (see also this MTF thread).
Advanced beer-spoiler detection medium (ABD) has been shown as a more effective growth medium compared to other media when attempting to grow beer-adapted bacteria such as hop tolerant strains of Lactobacillus that don't grow well on other media [5].
See also:
- Laboratory Techniques for the recipes for these agar types.
- Brewers Association list of lab services (filter by "Services", then "Contamination" or "Laboratory services").
- Kevin McGabe's HomebrewCon 2018 Seminar "Bringing a Brewery Quality Control Lab Into Your Home".
- MBAA guide to common spoilage bacteria and their morphology.
- MBAA TQ article, "Building a Quality Control Lab: An Introduction to Starting and Growing a Quality Program" by Melissa Antone.
- Review of Gram negative bacteria in brewing.
- Dr. Bryan Heit of Sui Generis blog explains his PCR method for detecting diastatic yeast.
- Zach Taggart offers QC tips on testing Lactobacillus growth using modified MRS agar plating and recording morphology.
- MBAA Podcast with Goose Island on how they traced an elusive wild yeast infection in their brewery.
- Books:
Viable But Nonculturable
"Viable but nonculturable" (VBNC) is a newly identified state for bacteria that are not able to grow or form colonies on typical growth media (i.e., lack of cell division), but they remain viable (alive) and retain a limited level of metabolic activity (reduced nutrient transport, respiration, and synthesis of compounds) while sometimes being able to regain their population when returned to a more ideal environment. The cells often exhibit dwarfing, and they can remain in this state without dying for 4-12+ months depending on the species. An appropriate viability test for a given species can be performed to show that the cells are not dead, even though they don't grow on typical growth media (for example, intracellular hydrolysis of CTC or reduction of INT as an indication of metabolic activity, by establishing the presence of an intact cytoplasmic membrane via BacLight® or propidium iodide, or by multi-parameter flow cytometry). Cells enter this state as a way to survive some sort of stress in their environment (for example, osmotic stress, too much oxygen exposure, exposure to white light, etc.). Treatments such as pasteurization in milk and chlorination of wastewater have also been shown to induce VBNC. A number of species have been found to be able to enter the VBNC state, including E. coli, Lactobacillus plantarum, L. lactis, L. linderi, L. casei, L. plantarum, L. paracollinocides, L. acetotolerans, and several species of Salmonella. Early studies on VBNC microbes were not able to fully show that the resuscitation was truly from VBNC cells rather than a very small number of culturable cells, but later studies were able to show that some bacteria can be resuscitated from a VBNC state, although most bacteria that enter a VBNC state have not been shown to be able to be resuscitated [68][69]. The concept of VBNC cells is somewhat controversial in microbiology; some experts argue that there is no difference between so-called "VBNC" cells and persister cells [70]. See also this MTF thread by Dr. Bryan Heit.
A couple of published studies have reported inducing the VBNC state in bacteria strains that were isolated from contaminated beer. Liu et al. (2017) were able to induce a VBNC state (meaning that they were not able to grow on growth media for up to 14 days) in a strain of Lactobacillus lindneri, a species that is responsible for 15-25% of spoiled beer reports, and determined VBNC via a Live/Dead BacLight® bacterial viability kit. They induced this state by storing the cells in beer at 0°C without shaking for 190 days. They also found that storing the VBNC cells at -80°C in glycerol stocks was the best way to maintain the cells. They were able to resuscitate the cells by growing them on MRS media that had 500-1000-μL of the enzyme catalase spread onto them (trying higher temperatures did not work to resuscitate, nor using higher concentrations of MRS), thus showing that brewers can use catalase to help grow VBNC state L. lindneri cells on MRS media (and perhaps other species of Lactobacillus as well). It took 3-4 days to begin showing signs of growth on the catalase supplemented MRS media. It was also demonstrated that VBNC cells could grow in beer after 30 days of incubation, and showed final cell counts similar to normal L. lindneri and resuscitated cells [71]. Lui et al. (2018) reproduced these results with a hop tolerant strain of L. brevis [69]. The genes that are associated with VBNC were also found in a beer contaminating strain of Lactobacillus acetolerans [72].
See also:
VBNC In Yeast
While the VBNC state has mostly only been studied in detail for bacteria, it has also been suggested that this state is also possible with eukaryotes (yeast). It has been reported that although there are many methods for detecting Brettanomyces in winemaking, there are cases when Brettanomyces is not found using culturing techniques, but years later have still infected wine. Agnolucci et al. (2010) found that sulfur dioxide induces the VBNC state in 7 Brettanomyces strains isolated from wine at concentrations of 0.2 mg/L (molecular SO2) for 5 out of the 7 strains to 0.4 mg/L for the other 2 strains after 24 hours of incubation in a synthetic wine medium that was supplemented with various amounts of sulfur dioxide. At 0.4 mg/L they found that all but one strain had 0% culturable cells, and 4-26% VBNC cells depending on the strain. At 0.8 mg/L, no strains had culturable cells, but they all had at least 4.6-17% VBNC cells (percent of the original number of cells before being exposed to the sulfur dioxide). Even at 1 mg/L of sulfur dioxide levels, there were 2.9-15% VBNC cells, depending on the strain (the ability for the Brettanomyces to remain viable at 1mg/L of sulfur dioxide might have also been due to the pH only being 3.5, and ethanol only being 13%). They also found that 2.1 mg/L was required to reduce the VBNC state of cells to zero after 55 days of incubation and limit the amount of ethyl phenols produced by Brettanomyces. They reported that trypan blue was the best method for detecting VBNC cells [73].
The Agnolucci et al. (2010) study did not provide a method or data for resuscitating the VBNC Brettanomyces cells so that they can again divide and grow colonies, and this has been criticized because resuscitation of VBNC cells is considered an important aspect of strengthening the conclusion that the cells are indeed VBNC. Serpaggi et al. (2012) found similar results with using 0.8 mg/L of molecular SO2, which resulted in all Brettanomyces cells from one wine strain to not be culturable on YPD after being incubated in synthetic wine medium that was supplemented with SO2 for 2 days, while the viability of the cells remained high for as long as 11 days (they did not check viability after 11 days, and the viability count remained constant from day 2 to day 11; viability was determined by staining with fluorescein diacetate which stains when certain metabolic esterase activity is present). They were able to resuscitate the cells by adding NaOH to the media to bring the pH up from 3.5 to 4.0 in order to effectively eliminate the molecular sulfur dioxide (molecular SO2 is the only form of SO2 that is significantly effective at inhibiting microbes, and it is only stable at very low pH's). They noted that VBNC cells were about 20% smaller in size than culturable cells. The cells in the VBNC state did not produce 4EG phenol but did produce a very small amount of 4EP phenol [74]. It has also been demonstrated that the presence of minerals and vitamins, as well as p-coumaric acid, can assist in resuscitating so-called VBNC cells of Brettanomyces [75].
It is worth noting that the VBNC state in Brettanomyces has not been tested against a higher concentration of SO2 (for example when wineries use much higher concentrations of SO2 solutions as a sanitizer), other chemical sanitizers, and pasteurization-level temperatures. For example, it has been proposed that steaming barrels in order for them to reach 140°F (60°C) for 20 minutes is enough to sanitize them (see Barrel Sanitizing for more information). However, Nunes de Lima et al. (2020) inoculated wines with various strains of Brettanomyces after raising the pH of the wine to 3.8, which rendered the amount of free SO2 in the wines completely ineffective, and many of the strains entered a VBNC state. This indicates that other unknown factors can induce a VBNC state in Brettanomyces. It has been documented that other factors, such as nutrient starvation, extreme temperatures, osmotic pressure and oxygen, has caused a VBNC state in other microorganisms (VBNC has been mostly studied in bacteria) [38].
Sensory Analysis
Sensory analysis is the process of implementing a standardized method for collecting data on finished beer through sensory evaluation. Sensory analysis can be a low tech approach to first steps in identifying potential quality issues [76].
See also:
- "Sensory Evaluation" on Craft beer & Brewing website.
- "An Introduction to Sensory Analysis" by Scott Bickham on the Morebeer website.
- BrewEssence product videos.
Supply Chain Management
Quality Improvement
(To do)
Quality improvement is a formal and systematic practice of analyzing ongoing processes and identifying problems or places for improvement.
See Also
Additional Articles on MTF Wiki
- Laboratory Techniques
- Wild Microbe Safety
- Mold
- Microbial Populations on Grain and Malted Barley
- Mixed Fermentation
External Resources
- Cleaning and Sanitation with Rick Theiner - BeerSmith Podcast #187.
- MBAA Podcast interview with Mary Pellettieri, author of "Quality Management: Essential Planning for Breweries".
- MBAA Podcast interview with Eric Jorgenson about his approach to microbiology and his quick reference guide of significant bacteria found in the brewery environment.
- MBAA Podcast interview with Lauryn Rivera and Tess Downer about Comprehensive Quality at Odell's.
- MBAA Podcast interview with Jack Van Paepeghem about troubleshooting and identifying Pectinatus (viscous and hydrogen sulfide in lagers) contamination in a CFT 10 head 2 seamer.
- "Quality Management: Essential Planning for Breweries" by Mary Pellettieri (Brewers Publications), 2015.
- "Illustrated Guide to Microbes and Sediments in Wine, Beer & Juice" by Charles G. Edwards (WineBuggs LLC), 2005 - A microscope companion book that includes over 30 different species of yeast, bacteria and mold commonly associated with beverages, as well as frequently encountered sediments.
- "QUALITY SYSTEMS: Practical Guides for Beer Quality", by Dr. Charlie Bamforth, 2019.
- "Brewery Cleaning: Equipment, Procedures, and Troubleshooting" by Richard J. Rench.
- Industry standards of dissolved oxygen levels in beer throughout the brewing process, by Hach.
- Eureka Brewing Blog on lab techniques, yeast handling, etc.
- "Yeast: A Practical Guide to Beer Fermentation" by Chris White and Jamil Zainesheff, 2010.
- "Brewing Yeast and Fermentation" by Boulton and Quain, 2006.
- American Society of Brewing Chemists.
- "Brewers' Laboratory Handbook: Brewing Without the Blindfold™", by Brewing Science Institute.
- "Selective Media Part 1" by BKYeast blog.
- "Pouring Plates and Making Slants" by BKYeast blog.
- Quality Management at Escarpment Labs.
- Teri Fahrendorf’s "Small Brewery Lab Procedures Manual"
- Colorado Brewers Guild "So You Want to Add a Brewing Lab?"
- Brew Strong podcast interview with Dr. Jon Hughes on Building A Quality Control Lab On A Budget (Aug 6, 2021).
- Bootleg Biology: a Semester-Long CURE Using Wild Yeast to Brew Beer.
References
- ↑ 1.0 1.1 "Quality Assurance vs. Quality Control". Diffen website. Retrieved 03/28/2018.
- ↑ Zachary Taggart. "Quality Assurance for Small Breweries: The Key to Growth". CBC 2018.
- ↑ Ciont, C.; Epuran, A.; Kerezsi, A.D.; Coldea, T.E.; Mudura, E.; Pasqualone, A.; Zhao, H.; Suharoschi, R.; Vriesekoop, F.; Pop, O.L. Beer Safety: New Challenges and Future Trends within Craft and Large-Scale Production. Foods 2022, 11, 2693. https://doi.org/10.3390/foods11172693.
- ↑ CONTAMINANT MICROBIOTA IN CRAFT BEERS. Marta García López, Elena Rocheb, Encarnación Rodríguez. 2020.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 125th Anniversary Review: Microbiological Instability of Beer Caused by Spoilage Bacteria. Ken Suzuki. 2012. DOI: https://doi.org/10.1002/j.2050-0416.2011.tb00454.x
- ↑ Distribution of Lactobacillus and Pediococcus in a Brewery Environment. Jorge Hugo Garcia-Garcia, Luis J. Galán-Wong, Benito Pereyra-Alférez, Luis C. Damas-Buenrostro, Esmeralda Pérez, and Juan Carlos Cabada. 2017. DOI: https://doi.org/10.1094/ASBCJ-2017-4294-01.
- ↑ 7.0 7.1 7.2 The Microbiology of Malting and Brewing. Nicholas A. Bokulich and Charles W. Bamforth. 2013. DOI: 10.1128/MMBR.00060-12
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 Enhancing the Microbiological Stability of Malt and Beer — A Review. Anne Vaughan, Tadhg O'Sullivan, Douwe Van Sinderen. 2005. DOI: https://doi.org/10.1002/j.2050-0416.2005.tb00221.x.
- ↑ Comparative genetic and physiological characterisation of Pectinatus species reveals shared tolerance to beer-associated stressors but halotolerance specific to pickle-associated strains. Timo Kramer, Philip Kelleher, Julia van der Meer, Tadhg O’Sullivan, Jan-Maarten A.Geertman, Sylvia H. Duncan, Harry J. Flint, Petra Louis. 2020. DOI: https://doi.org/10.1016/j.fm.2020.103462.
- ↑ Beer‐spoilage characteristics of Staphylococcus xylosus newly isolated from craft beer and its potential to influence beer quality. Zhimin Yu, Qiuying Luo, Li Xiao, Yumei Sun, Rong Li, Zhen Sun, Xianzhen Li. 2019. DOI: 10.1002/fsn3.1256.
- ↑ 11.00 11.01 11.02 11.03 11.04 11.05 11.06 11.07 11.08 11.09 11.10 Disinfectant testing against brewery-related biofilms. Erna Storgårds, Gun Wirtanen. 2001.
- ↑ 12.0 12.1 A Review of Molecular Methods for Microbial Community Profiling of Beer and Wine. Nicholas A. Bokulich, Charles W. Bamforth & David A. Mills. 2018.
- ↑ 13.0 13.1 13.2 13.3 13.4 13.5 Process hygiene control in beer production and dispensing. Erna Storgårds. VTT Publications 410. 2000.
- ↑ Alex R. Thompson, Julie K. Northcutt and Paul Dawson. Bacterial contamination and surface hygiene in the microbrewery environment. Journal of Brewing and Distilling. Vol. 13(1), pp. 1-10, January-June 2024. DOI: 10.5897/JBD2024.0060.
- ↑ 15.0 15.1 15.2 15.3 15.4 Mapping microbial ecosystems and spoilage-gene flow in breweries highlights patterns of contamination and resistance. Nicholas A Bokulich, Jordyn Bergsveinson, Barry Ziola, David A Mills. 2015.
- ↑ The Paradox of Mixed‐Species Biofilms in the Context of Food Safety. Iqbal Kabir Jahid and Sang‐Do Ha. 2014.
- ↑ Biofilms in the Food and Beverage Industries. P M Fratamico, B A Annous, N W Guenther. Elsevier, Sep 22, 2009. Pp 4-14.
- ↑ The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Shou-Fu Duan, Pei-Jie Han, Qi-Ming Wang, Wan-Qiu Liu, Jun-Yan Shi, Kuan Li, Xiao-Ling Zhang & Feng-Yan Bai. 2018.
- ↑ Bakers' yeast, a model for fungal biofilm formation. Reynolds and Fink. 2011. DOI: 10.1126/science.291.5505.878.
- ↑ 20.0 20.1 Microbial attachment and biofilm formation in brewery bottling plants. Erna Storgårds, Kaisa Tapani, Peter Hartwall, Riitta Saleva & Maija-Liisa Suihko. 2006. DOI: https://doi.org/10.1094/ASBCJ-64-0008.
- ↑ Disinfection in Food Processing – Efficacy Testing of Disinfectants. G. Wirtanen, S. Salo. 2003.
- ↑ "What are molds?". CDC website. Retrieved 01/25/2019.
- ↑ Differences and Similarities Among Proteolytic and Nonproteolytic Strains of Clostridium botulinum Types A, B, E and F: A Review. RICHARD K. LYNT*, DONALD A. KAUTTER and HAIM M. SOLOMON. 1982.
- ↑ Chris Colby. "Storing Wort Runs the Risk of Botulism". Beer and Wine Journal Blog. 04/17/2014. Retrieved 04/04/2018.
- ↑ Bryan Heit. Milk The Funk Facebook thread on yeast and bacteria spores and brewery hygiene. 04/04/2018.
- ↑ Quain, D. E. (2021) The enhanced susceptibility of alcohol-free and low alcohol beers to microbiological spoilage: implications for draught dispense, J. Inst. Brew., XXX, doi: https://doi.org/10.1002/jib.670.
- ↑ Caroline Smith from Lallemand. Milk The Funk Facebook group post on diastaticus contamination. Feb 2018.
- ↑ Modeling the inactivation of Lactobacillus brevis DSM 6235 and retaining the viability of brewing pitching yeast submitted to acid and chlorine washing. Munford, A.R.G., Chaves, R.D., Granato, D. et al. Appl Microbiol Biotechnol (2020). https://doi.org/10.1007/s00253-020-10534-x.
- ↑ "Lysozyme". Wikipedia. Retrieved 0319/2020.
- ↑ 2018 Fermentation Handbook. Scott Laboratories. Retrieved 03/19/2020.
- ↑ Lysozyme in Wine: An Overview of Current and Future Applications. Marco Esti, Ilaria Benucci. Comprehensive Reviews in Food Science and Food Safety. 2014.
- ↑ Van Landschoot, Anita & Villa, A. (2005). Antibacterial properties of hen egg white lysozyme against beer spoilage bacteria and effect of lysozyme on yeast fermentation.
- ↑ Müller-Auffermann, K, Grijalva, F, Jacob, F, and Hutzler, M (2015), Nisin and its usage in breweries: a review and discussion. J. Inst. Brew., 121, 309–319. doi: 10.1002/jib.233.
- ↑ "Sloppy Slurry vs. Clean Starter". Brulosophy website. 2015. Retrieved 03/19/2020.
- ↑ 35.0 35.1 Gerhard J. Haas. "Microbial Control Methods in the Brewery". Editor(s): Wayne W. Umbreit. Advances in Applied Microbiology. Academic Press, Volume 2, 1960. Pages 113-162. ISSN 0065-2164. ISBN 9780120026029.
- ↑ THERMAL DEATH POINTS OF MICRO-ORGANISMS IN BEER. Aage Lund. 1947.
- ↑ Thermal inactivation of the wine spoilage yeasts Dekkera/Brettanomyces. José António Couto, Filipe Neves, Francisco Campos, Tim Hogg. 2005. DOI: 10.1016/j.ijfoodmicro.2005.03.014.
- ↑ 38.0 38.1 Survival and metabolism of hydroxycinnamic acids by Dekkera bruxellensis in monovarietal wines. Adriana Nunes de Lima, Rui Magalhães, Francisco Manuel Campos, José António Couto. 2020. DOI: https://doi.org/10.1016/j.fm.2020.103617.
- ↑ Microfiltration for Filtration and Pasteurization of Beers. Guilherme dos Santos Bernardi, Jacir Dal Magro, Marcio A. Mazutti, J. Vladimir Oliveira, Marco Di Luccio, Giovani Leone Zabot, Marcus V. Tres. 2019. DOI: https://doi.org/10.1016/B978-0-12-815258-4.00013-5.
- ↑ Spoilage yeasts in beer and beer products. Inge M Suiker, Han AB Wösten. Current Opinion in Food Science. Microbiology, Department of Biology, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands. 02/19/2022.
- ↑ Brandon Jones. Private correspondence with Dan Pixley. 04/02/2018.
- ↑ Levader on Reddit.com. "The Brewery". Retrieved 04/02/2018.
- ↑ Susceptibility of wine spoilage yeasts and bacteria in the planktonic state and in biofilms to disinfectants. Mariana Tristezza, António Lourenço, André Barata, Luísa Brito, Manuel Malfeito-Ferreira, Virgílio Loureiro. 2010.
- ↑ Adhesion and biofilm production by wine isolates of Brettanomyces bruxellensis. C. M. Lucy Joseph, Gagandeep Renuka Kumar, Gagandeep Renuka Kumar, Edward Su, Linda F Bisson. 2006. American Journal of Enology and Viticulture 58(3):373-378.
- ↑ Disinfectant testing against brewery-related biofilms. Storgårds, Erna & Närhi, Mikko & Wirtanen, Gun. 2001.
- ↑ COMMERCIAL SANITIZERS EFFICACY – A WINERY TRIAL. Duarte, Filomena & López, Alberto & Alemão, Filomena & Santos, Rodrigo & Canas, Sara. 2011.
- ↑ Private correspondence with Dr. Bryan Heit by Dan Pixley. 04/12/2018.
- ↑ Dr. Bryan Heit. Milk The Funk Facebook thread on an MBAA podcast about the efficacy of iodophor as a sanitizer. 07/10/2018.
- ↑ Dr. Molly Kelly. "Understanding Difficult Malolactic Fermentations". Wine & Grapes U. 10/26/2018. Retrieved 11/22/2018.
- ↑ Brettanomyces bruxellensis yeasts: impact on wine and winemaking. Monica Agnolucci, Antonio Tirelli, Luca Cocolin, Annita Toffanin. 2017.
- ↑ "How to Prevent Glass Corrosion". Paul F. Duffer. Glass Digest, November 15, 1986.
- ↑ Shawn Savuto. Milk The Funk Facebook group post on caustic erroding glass. 08/12/2018.
- ↑ 53.0 53.1 Removal of Microbial Biofilms from Dispense Equipment: The Effect of Enzymatic Pre-digestion and Detergent Treatment. Samantha L. Walker, Michail Fourgialakis, Belen Cerezo, and Stephen Livens. 2007.
- ↑ User 'S. cerevisiae'. American Homebrewers Association forums. 10/05/2015. Retrieved 04/11/2018.
- ↑ User 'richardt'. American Homebrewers Association forums. 11/15/2010. Retrieved 04/11/2018.
- ↑ Coopers Community Forums. Post by user 'Christinas1'. 09/08/2016. Retrieved 05/23/2019.
- ↑ Conn, Denny. American Homebrew Association forums. 04/08/2018. retrieved 07/09/2018.
- ↑ Efficacy of Two Acidic Sanitizers for Microbial Reduction on Metal Cans and Low-Density Polyethylene Film Surfaces. J. LEE, M.J. GUPTA, J. LOPES, AND M.A. PASCALL. 2007.
- ↑ Minimum inhibitory concentrations of antimicrobials against micro-organisms related to citrus juice. P.P Winniczuk, M.E Parish. 1997.
- ↑ Dr. Bryan Heit. Milk The Funk Facebook thread on an MBAA podcast about the efficacy of iodohor as a sanitizer. 07/10/2018.
- ↑ "Non-caustic composition comprising peroxygen compound and metasilicate and cleaning methods for using same". US patent for PBW. 03/01/1995. Retrieved 04/19/2018.
- ↑ "PBW Tech Sheet". Five Star Chemicals. Retrieved 04/19/2018.
- ↑ Homebrewtalk thread reporting PBW breaking down PET bottles. 03/25/2013.
- ↑ Better Bottle Cleaning and Sanitizing web article; downloaded from Archive.org.
- ↑ Disposable electrochemical biosensors for Brettanomyces bruxellensis and total yeast content in wine based on core-shell magnetic nanoparticles. María L. Villalong, Boryana Borisova, Christian B. Arenas, Anabel Villalonga, María Arévalo-Villena, Alfredo Sánchez, José M. Pingarrón, Ana Briones-Pérez, Reynaldo Villalong. 2018.
- ↑ Nick Impellitteri. Milk The Funk Facebook post on the ability of different media to grow beer adapted Brettanomyces. 04/03/2018.
- ↑ Effects of Beer Adaptation on Culturability of Beer-Spoilage Dekkera/Brettanomyces Yeasts. Koji Suzuki, Shizuka Asano, Kazumaru Iijima, Tomoo Ogata, Yasushi Kitagawa & Tsunehiro Ikeda. 2018.
- ↑ The Viable but Nonculturable State in Bacteria. James D. Oliver. The Journal of Microbiology. 2005.
- ↑ 69.0 69.1 Induction and Recovery of the Viable but Nonculturable State of Hop-Resistance Lactobacillus brevis. Junyan Liu, Yang Deng, Thanapop Soteyome, Yanyan Li, Jianyu Su, Lin Li, Bing Li, Mark E. Shirtliff, Zhenbo Xu, and Brian M. Peters. Front. Microbiol. 2018. DOI: https://doi.org/10.3389/fmicb.2018.02076.
- ↑ Song, S. and Wood, T.K. (2021), ‘Viable but non-culturable cells’ are dead. Environ Microbiol, 23: 2335-2338. https://doi.org/10.1111/1462-2920.15463.
- ↑ First study on the formation and resuscitation of viable but nonculturable state and beer spoilage capability of Lactobacillus lindneri. Junyan Liu, Lin Lia, Bing Li, Brian M. Peters, Yang Deng, Zhenbo Xua, Mark E. Shirtliff. Microbial Pathogenesis, Vol 107. 2017. DOI: https://doi.org/10.1016/j.micpath.2017.03.043.
- ↑ Chunguang Luan, Weihua Cao, Na Luo, Jingxia Tu, Jianqin Hao, Yihong Bao, Feike Hao, Deliang Wang & Xin Jiang (2021) Genomic Insights into the Adaptability of the Spoilage Bacterium Lactobacillus acetotolerans CN247 to the Beer Microenvironment, Journal of the American Society of Brewing Chemists, DOI: 10.1080/03610470.2021.1997280.
- ↑ Sulphur dioxide affects culturability and volatile phenol production by Brettanomyces/Dekkera bruxellensis. Agnolucci M, Rea F, Sbrana C, Cristani C, Fracassetti D, Tirelli A, Nuti M. 2010. DOI: https://doi.org/10.1016/j.ijfoodmicro.2010.07.022.
- ↑ Characterization of the "viable but nonculturable" (VBNC) state in the wine spoilage yeast Brettanomyces. Serpaggi V, Remize F, Recorbet G, Gaudot-Dumas E, Sequeira-Le Grand A, Alexandre H. 2012. DOI: 10.1016/j.fm.2011.12.020.
- ↑ Chandra M, Branco P, Prista C, Malfeito-Ferreira M. Role of p-Coumaric Acid and Micronutrients in Sulfur Dioxide Tolerance in Brettanomyces bruxellensis. Beverages. 2023; 9(3):69. https://doi.org/10.3390/beverages9030069.
- ↑ "Sensory Evaluation" Craft Beer & Brewing website. Retrieved 02/14/2020.